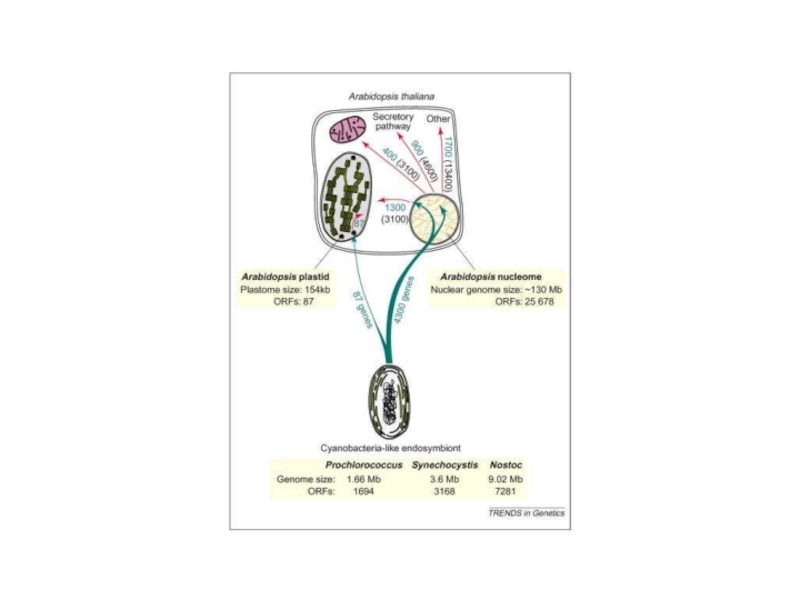
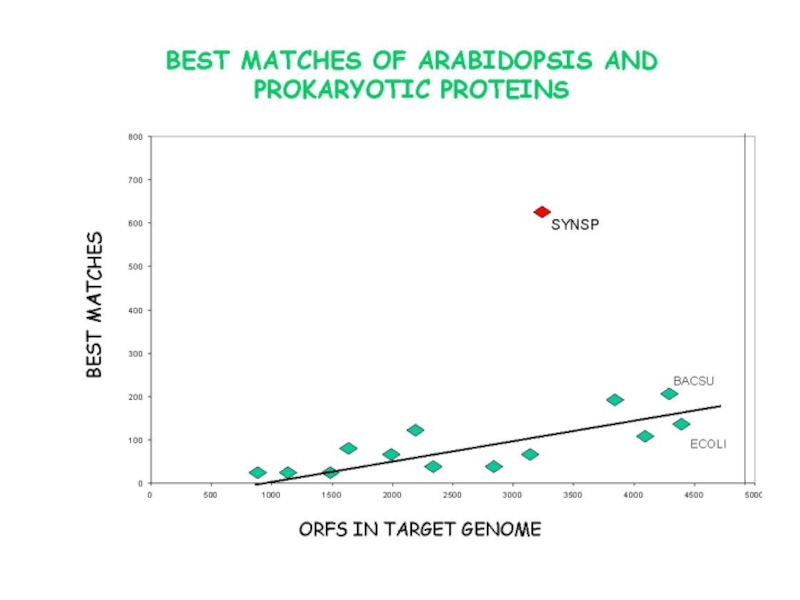
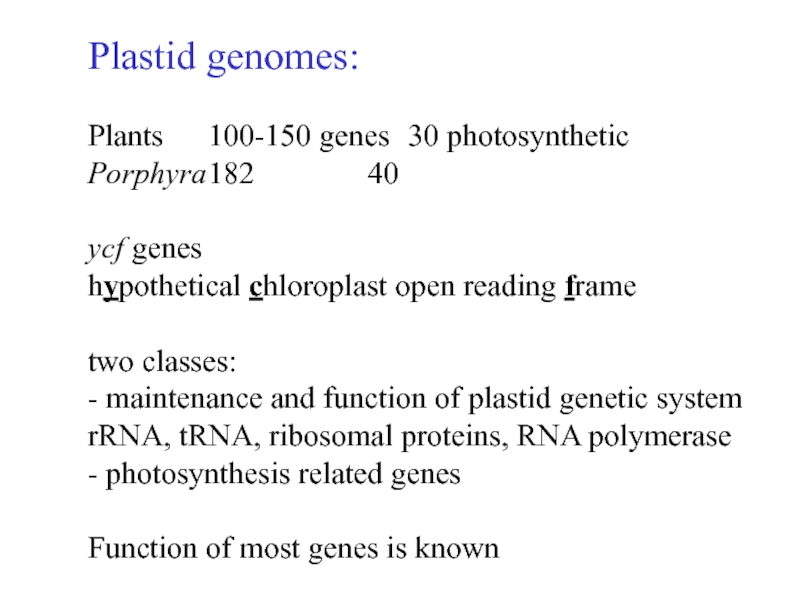
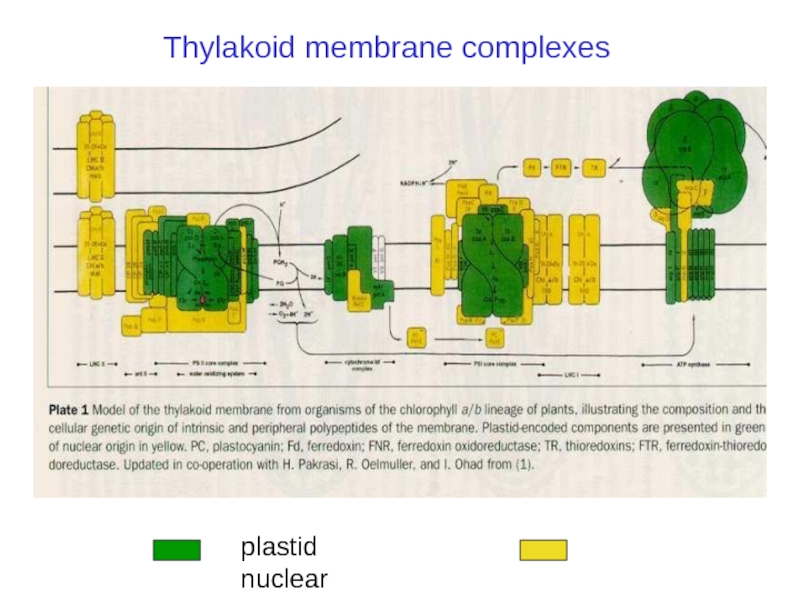
Слайд 5Plastid genomes:
Plants 100-150 genes 30 photosynthetic
Porphyra 182 40
ycf genes
hypothetical chloroplast open reading frame
two
classes:
- maintenance and function of plastid genetic system
rRNA, tRNA, ribosomal
proteins, RNA polymerase
- photosynthesis related genes
Function of most genes is known
Слайд 8Copyright ©2002 by the National Academy of Sciences
Martin, William et
al. (2002) Proc. Natl. Acad. Sci. USA 99, 12246-12251
Phylogeny of
chloroplast genomes, gene loss, and gene transfer
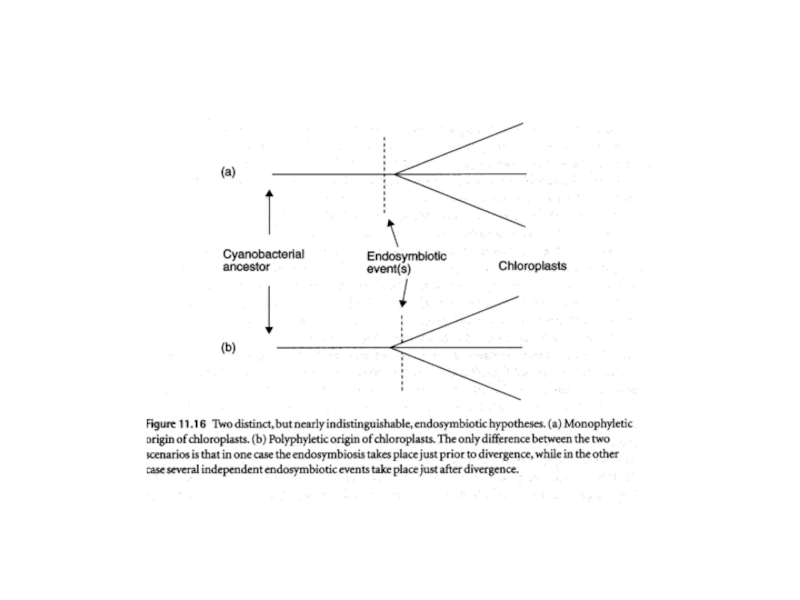
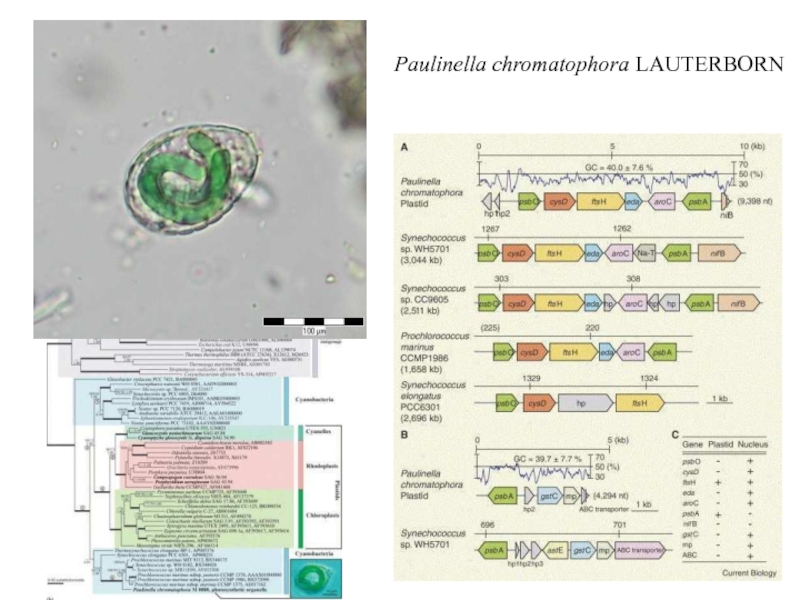
Слайд 12Fig. 3. Endosymbiosis in the history of plastid evolution. All
primary (top), secondary (middle), serial secondary and tertiary (bottom) endosymbiotic
associations mentioned in the text are represented here. Cells are color-coded so that the cytoplasmic color matches the color of the supergroup in Fig. 1 to which that eukaryote belongs (Cercozoa are yellow, plants are green, excavates are purple, and chromalveolates are blue). Plastids are color-coded to distinguish the three primary plastid lineages (cyanobacteria and glaucophyte plastids are both blue-green, red algal plastids are red, and green algal plastids are dark green). Primary endosymbiosis: At the top left, the cercozoan euglyphid amoeba Paulinella takes up a Synechococcus-like cyanobacterium and retains two apparently permanent endosymbionts, losing its feeding pseudopods. This may represent an independent primary endosymbiosis. At the top center, a cyano bacterium of unknown type is taken up by an ancestor of the plant supergroup, the direct descendent of which are the three primary algal lineages, glaucophytes, red algae, and green algae. Glaucophytes and red algae retain phycobilisomes, and glaucophytes retain the peptidoglycan wall. Plants are derived from green algae. Secondary endosymbiosis: At the center right, two green algae are independently taken up by two eukaryotes, one cercozoan (yellow) and one excavate (purple) giving rise to the chlorarachniophytes and euglenids, respectively. Euglenids have three-membrane plastids, and chlorarachniophytes retain a nucleomorph. At the center, a red alga is taken up by an ancestor of the chromalveolates (light blue), giving rise to cryptomonads, haptophytes, heterokonts, and alveolates (dinoflagellates, apicomplexa, and ciliates). In cryptomonads, haptophytes, and heterokonts, the outer membrane of the plastid is continuous with the rough ER and nuclear envelope, and cryptomonads also retain a nucleomorph and phycobilisomes (which are inside the thylakoid lumen rather than on the outer surface). The presence of a plastid in ciliates is purely conjectural at present, and there is no direct evidence for this organelle. Dinoflagellates have a three-membrane plastid (the peridinin-containing plastid) that has been replaced on several occasions by serial secondary and tertiary endosymbiosis: At bottom right, a green alga is taken up by a dinoflagellate in a serial secondary endosymbiosis giving rise to Lepidodinium and close relatives. At bottom left, three different dinoflagellates take up a cryptomonad, a haptophyte, and a diatom, giving rise to Dinophysis, Karenia, and Kryptoperidinium, respectively. Each of these plastids has lost one or more membranes, and how plastid targeting works is completely unknown. Kryproperidinium retains the diatom nucleus and also a three-membrane eyespot, suggested to be the ancestral plastid