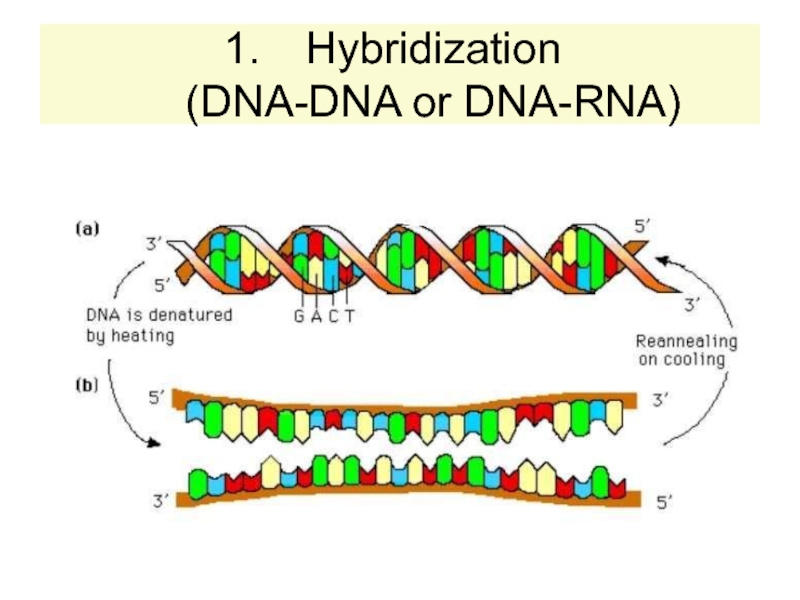
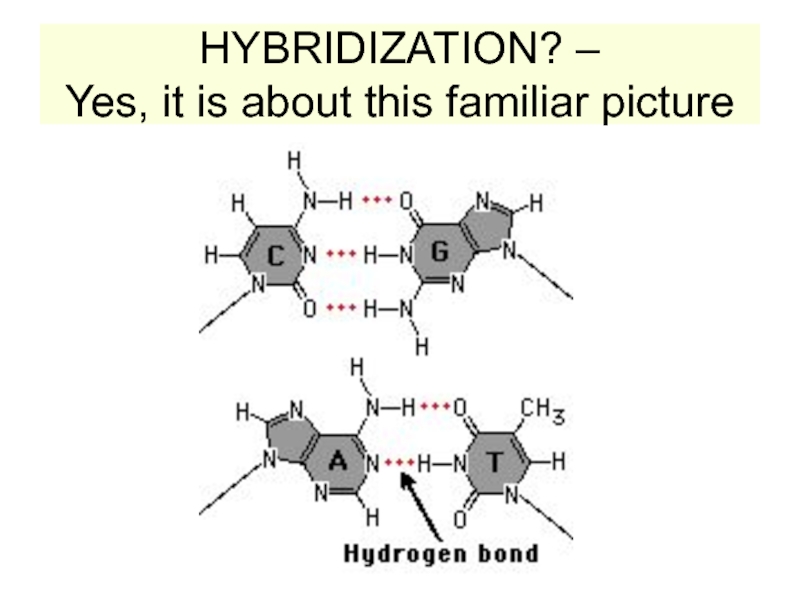
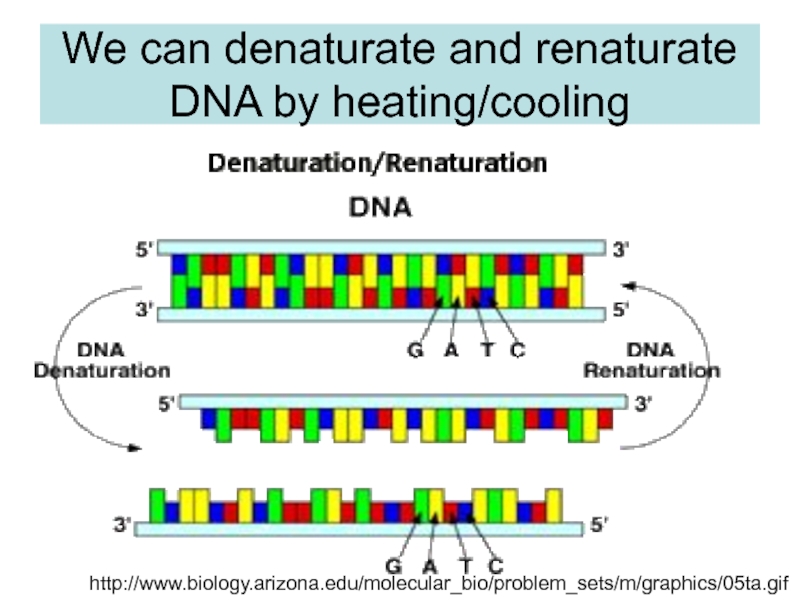
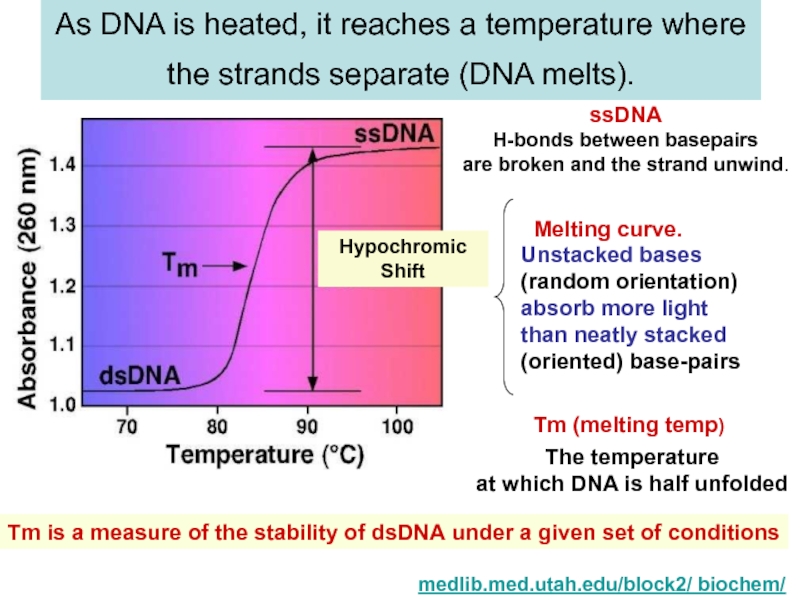
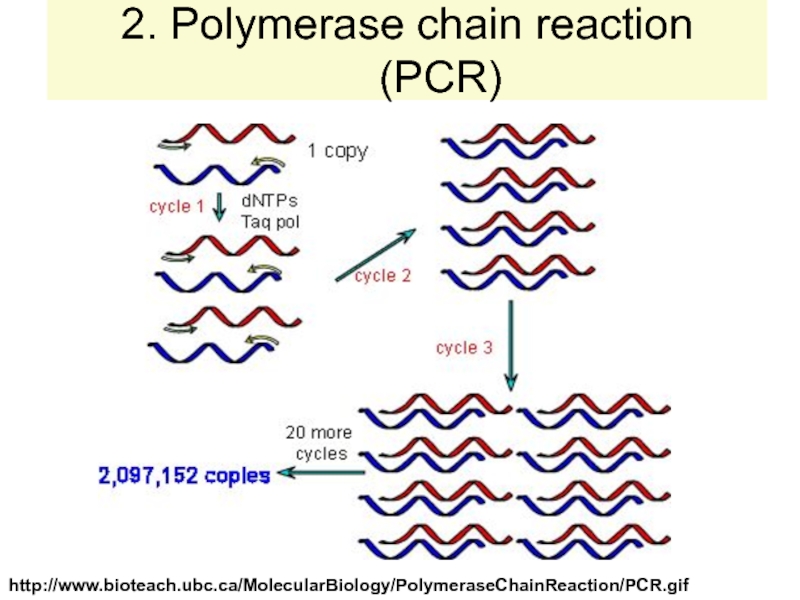
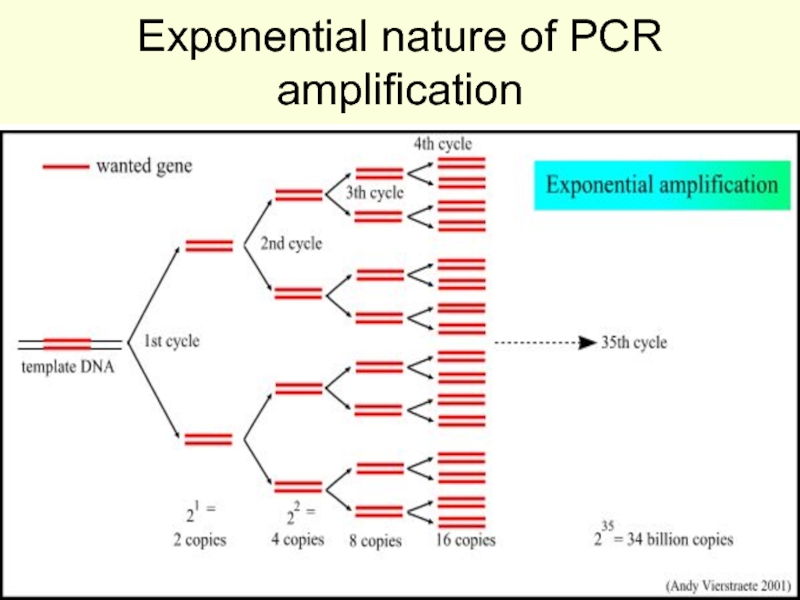
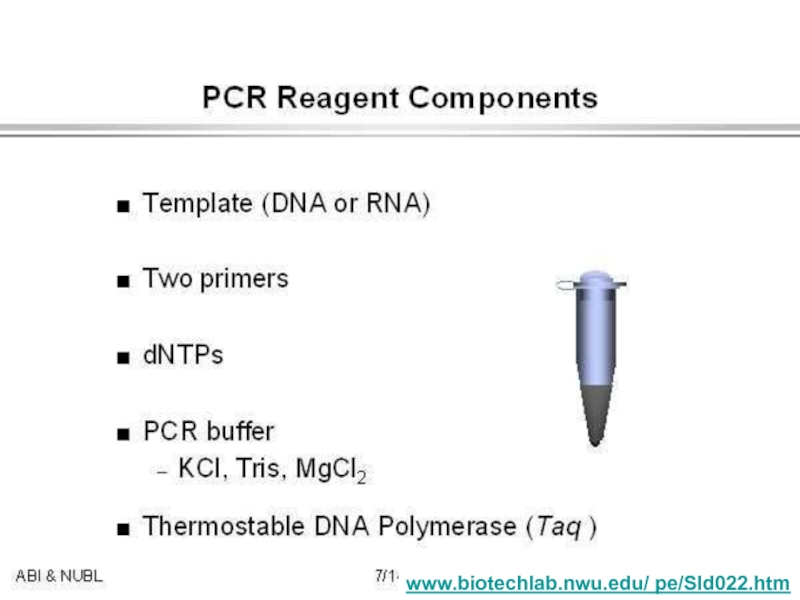
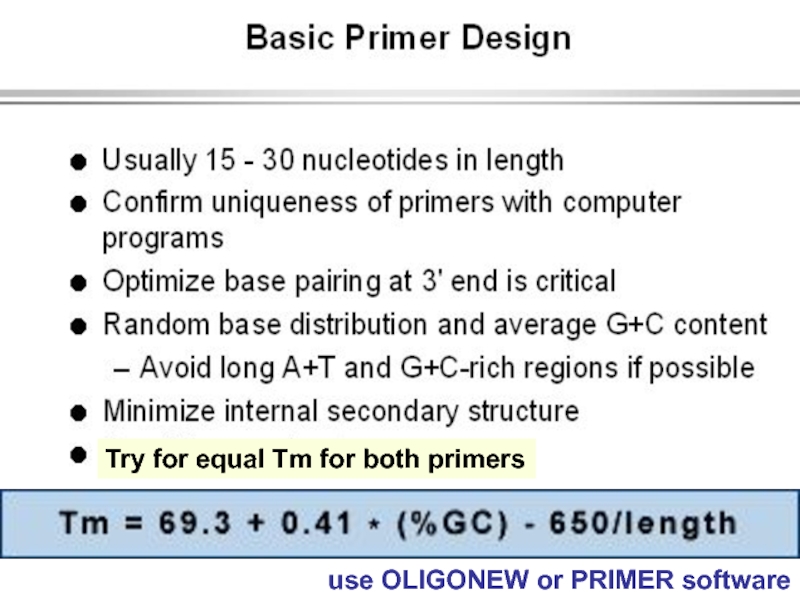
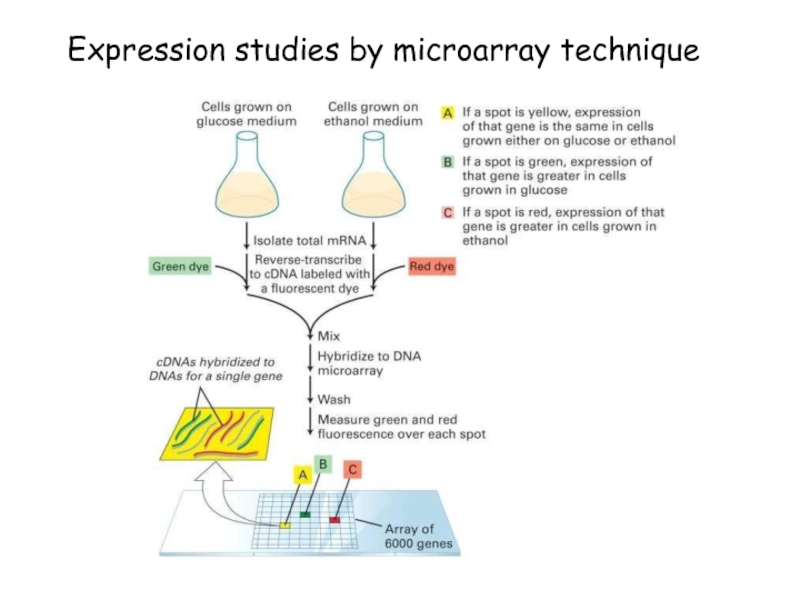
strands separate (DNA melts).
medlib.med.utah.edu/block2/ biochem/
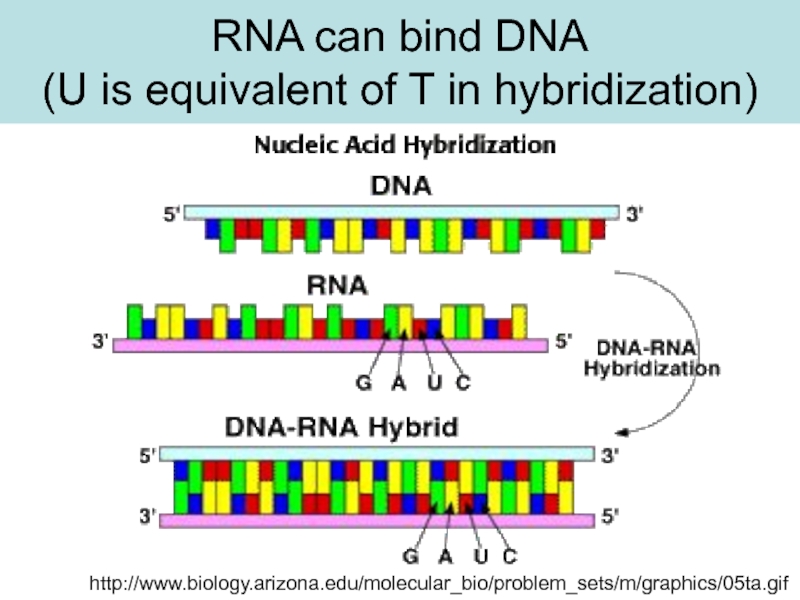
ssDNA
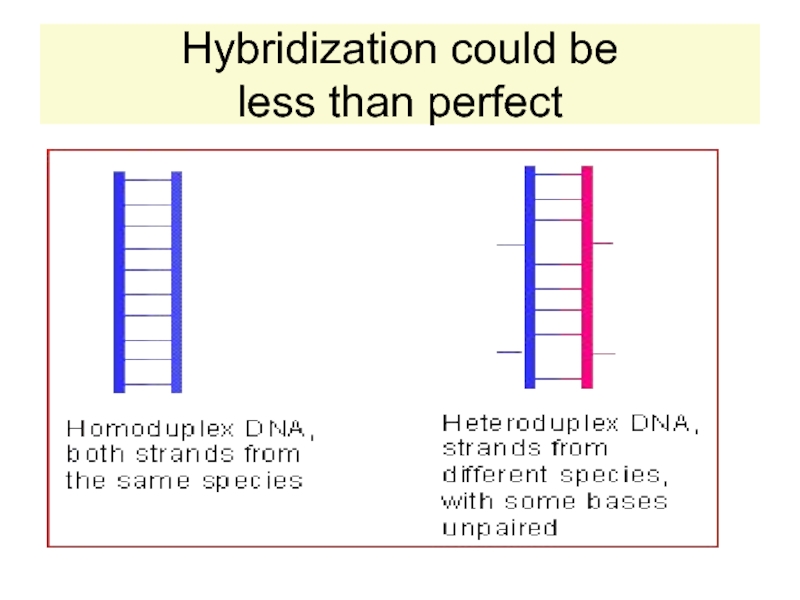
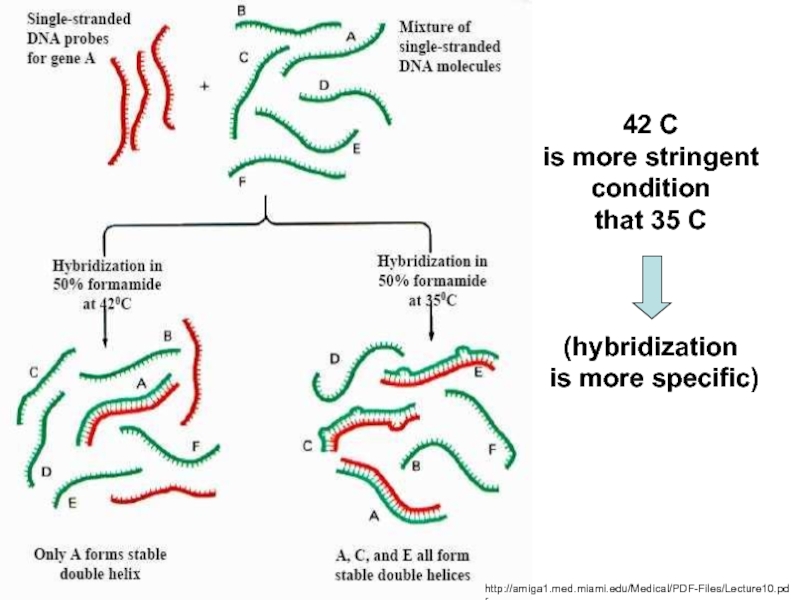
H-bonds between basepairs
are broken and the strand unwind.
Unstacked bases
(random orientation)
absorb more light
than neatly stacked
(oriented) base-pairs
Melting curve.
The temperature
at which DNA is half unfolded
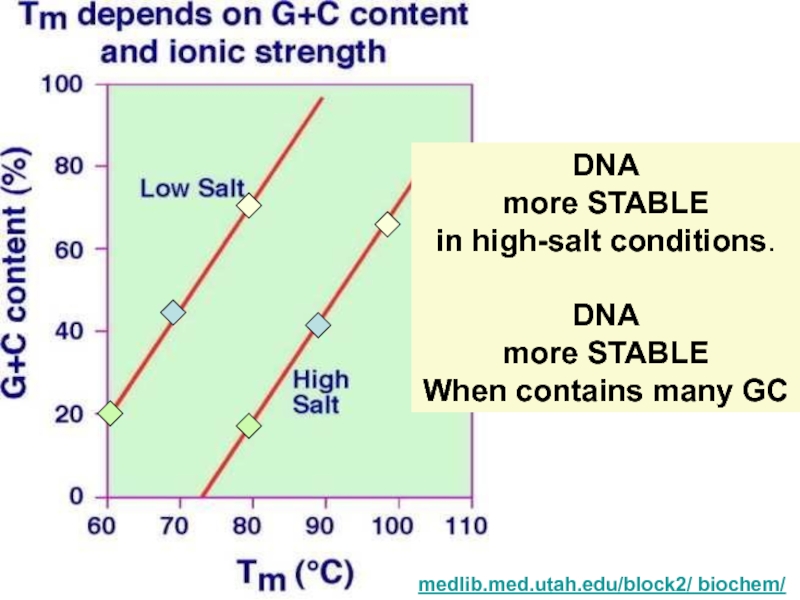
Tm (melting temp)
Tm is a measure of the stability of dsDNA under a given set of conditions
Hypochromic
Shift