Слайд 1 Lactococcus sp.
Done by:
Naizabayeva D.
BT 16-02
Слайд 2Content
Introduction (preface)
History and discovery
Taxonomy
Biology (Morphological, physiological and biochemical
features)
Phenotypic (morphological and physiologic) analysis
Immunological (serological analysis)
Genotypic techniques
Manipulation (genetic
or non-genetic)
Facts
References
Слайд 3Introduction
Lactic acid bacteria are among the most important groups
of microorganisms used in food fermentations. They contribute to the
taste and texture of fermented products and inhibit food spoilage bacteria by producing growth-inhibiting substances and large amounts of lactic acid. As agents of fermentation LAB are involved in making yogurt, cheese, cultured butter, sour cream, sausage, cucumber pickles, olives and sauerkraut, but some species may spoil beer, wine and processed meats.
Meanwhile, they has a great application in sphere of pharmacy, due to their antimicrobial ability and safety for the human, taken a probiotics, produce practically important bactericines , and recently practiced as a delivery agent for therapeutics and vaccines.
Слайд 42.History and discovery
The common organism associated with the souring of
milk was first described by Lister (1878) as 'Bacterium lactis'.
In the next thirty years a variety of names and descriptions, including Str. lacticus of Kruse (1903), were added to the literature, and it was left to Lohnis (1909) to clarify the situation. He agreed that it should be placed in the genus Streptococcus and suggested that it should be named 'Streptococcus lactis', thus keeping Lister's original species name. The various synonyms encountered are very numerous and were summarized by Breed (1928). Bergey (1939) has adopted this list in its entirety. Orla Jensen's (1919) description of Str. lactis is as follows. The optimum temperature for growth is 30°C., and when freshly isolated it will coagulate sterile milk at this temperature in less than 24 hr. At the optimum temperature it occurs as a diplococcus or as short chains. Growth below 10 °C. and above 40°C is in general poor. Str. lactis is characterized by failure to ferment sucrose.
Слайд 52.History and discovery
This group of bacteria, previously designated the lactic
streptococci (Streptococcus lactis subsp. lactis or S. lactis subsp. cremoris) was placed in this new taxon
in 1985 by Schleifer. This discovery were investigated according to results of nucleic acid hybridization studies and immunological relationships of superoxide dismutase, which in its turn demonstrated that Streptococcus lactis (and its subspecies), Lactobacillus xylosus, Lactobacillus hordniae, S. garvieae, S. plantarum and S. raffinolactis are closely related to each other but not to other streptococci. Therefore it was proposed that these taxa be transferred to a new genus Lactococcus gen.nov. as Lactococcus lactis subsp. lactis (including former S. lactis subsp. diacetilactis and Lactobacillus xylosus) comb.nov., L.lactis subsp. cremoris comb.nov., L.actis subsp. hordniae comb.nov., L.garvieae comb.nov., L.plantarum comb.nov. and L.raffinolactis comb.nov. The relatedness of these organisms has also been demonstrated by the similarity of their lipoteichoic acid structures, lipid pattern, fatty acid and menaquinone compositions.

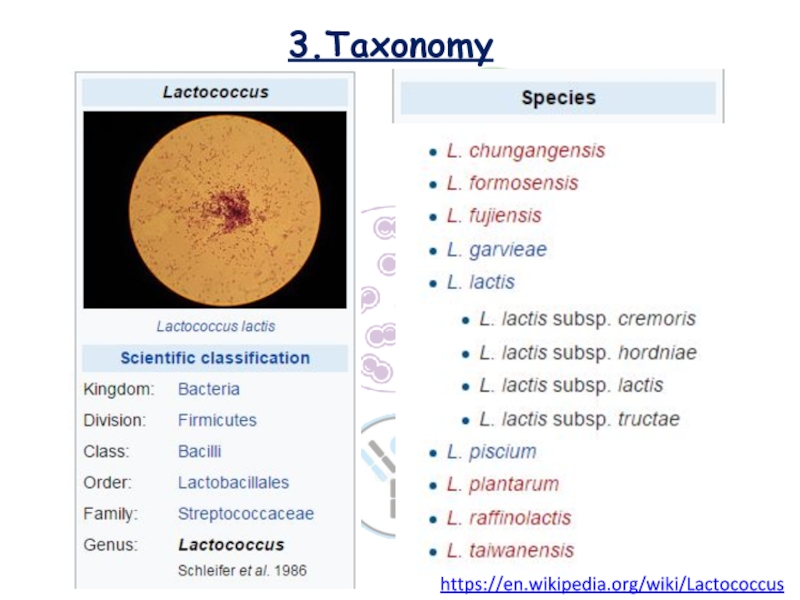
Слайд 73.Taxonomy
https://en.wikipedia.org/wiki/Lactococcus
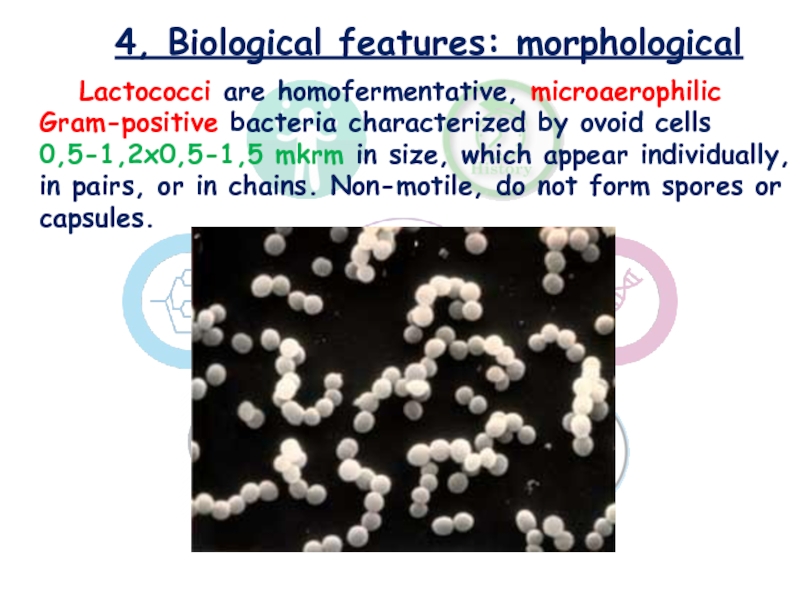
Слайд 84, Biological features: morphological
Lactococci are homofermentative, microaerophilic Gram-positive bacteria characterized
by ovoid cells 0,5-1,2x0,5-1,5 mkrm in size, which appear individually,
in pairs, or in chains. Non-motile, do not form spores or capsules.
Слайд 94, Biological features: physiological
http://textbookofbacteriology.net/lactics.html
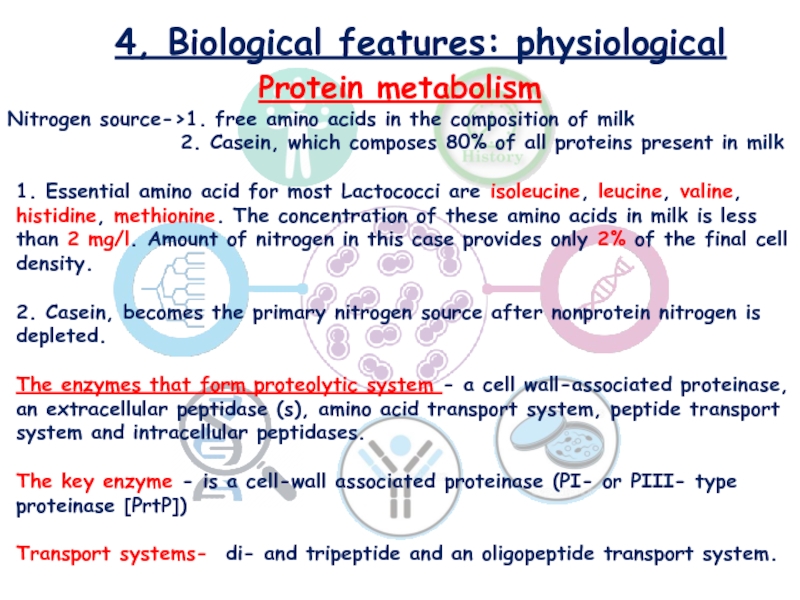
Слайд 104, Biological features: physiological
Protein metabolism
Nitrogen source->1. free amino acids
in the composition of milk
2. Casein, which composes 80% of all proteins present in milk
1. Essential amino acid for most Lactococci are isoleucine, leucine, valine, histidine, methionine. The concentration of these amino acids in milk is less than 2 mg/l. Amount of nitrogen in this case provides only 2% of the final cell density.
2. Casein, becomes the primary nitrogen source after nonprotein nitrogen is depleted.
The enzymes that form proteolytic system - a cell wall-associated proteinase, an extracellular peptidase (s), amino acid transport system, peptide transport system and intracellular peptidases.
The key enzyme - is a cell-wall associated proteinase (PI- or PIII- type proteinase [PrtP])
Transport systems- di- and tripeptide and an oligopeptide transport system.
Слайд 114, Biological features: physiological
Strains: 1, CAU 28T;
2, L. garvieae
KCTC 3772T;
3, L. piscium DSM
6634T;
4, L. plantarum DSM
20686T;
5, L. raffinolactis DSM 20443T;
6,L. lactis subsp. cremoris KCCM 40699T;
7, L. lactis subsp. hordniae
KCTC 3768T;
8, L. lactis subsp. lactis KCTC 3769T.
(+) Positive,(-) negative,
w- weakly positive
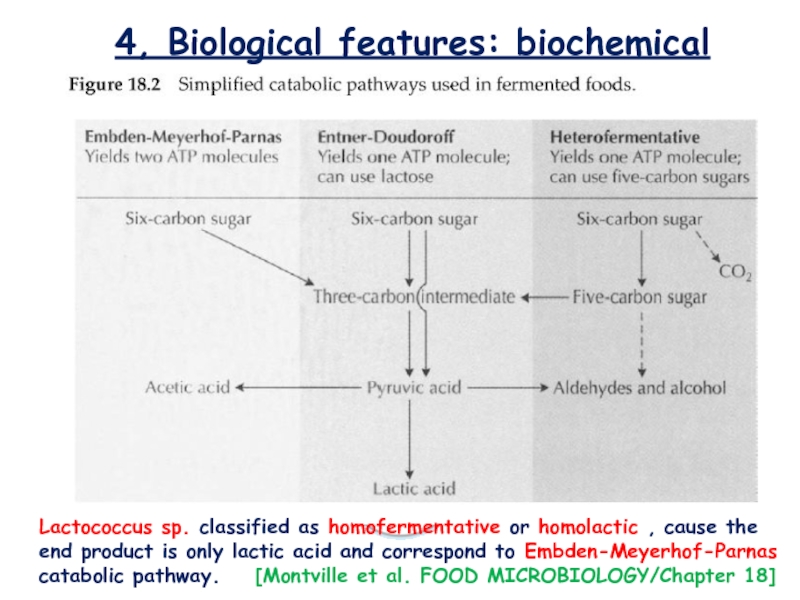
Слайд 124, Biological features: biochemical
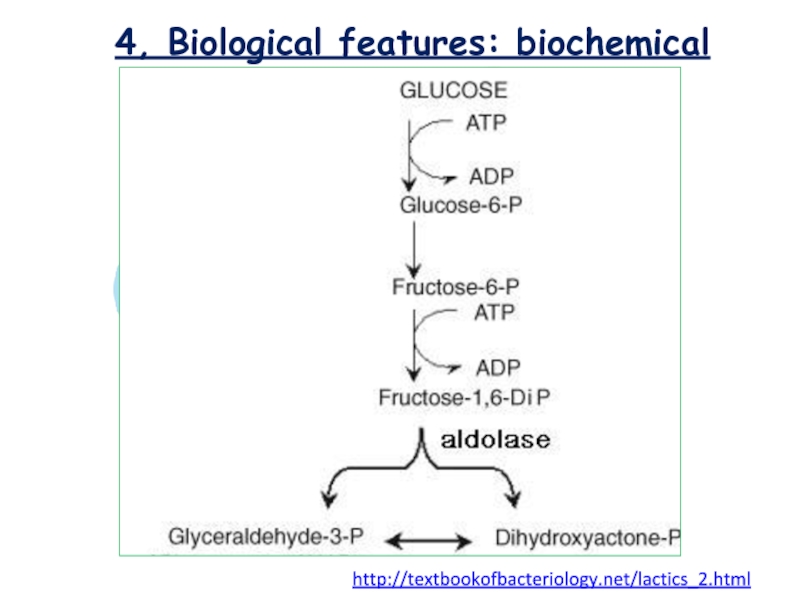
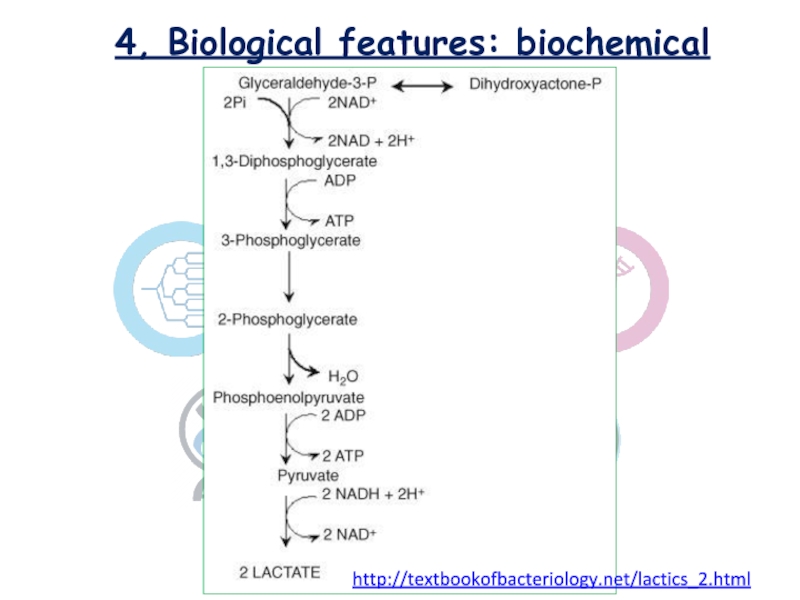
Lactococcus sp. classified as homofermentative or homolactic
, cause the end product is only lactic acid and
correspond to Embden-Meyerhof-Parnas catabolic pathway. [Montville et al. FOOD MICROBIOLOGY/Chapter 18]
Слайд 134, Biological features: biochemical
http://textbookofbacteriology.net/lactics_2.html
Слайд 144, Biological features: biochemical
http://textbookofbacteriology.net/lactics_2.html
Слайд 154, Biological features: biochemical
Capability to metsbolize citrate
Only Lc. lactis subsp.
lactis biovar diacetylactis has the ability to metabolize the citrate
present in milk.
Citrate
Oxaloacetate
Pyruvate
acetaldehyde-thiaminopyrophosphate (TPP)
α-acetolactate
acetoin
Decarboxylation reactions
Total endproducts:
Acetic acid, diacetyl,
Acetoin, 2,3 butanediol, CO2, which contributes flavor development of fermented food
Catalysed by the citrat permease (CitP) encoded in Plasmid-citP, operon-citQRP
Слайд 16Detection and analysis
1. Morphological
2. Physiological / biochemical
3. Serological
4.
Genetic
Microscopy (Gram staining )
Medium associated tests (antibiotic resistance, carbohydrate
fermentation, acetoin production and etc. ), API 50CHL strips (fermentation of 49 carbohydrates and esculin hydrolysis), Biolog plate (96 carbohydrates)
Slide agglutination, fluorescent
antibody staining, flow-cytometry based methods
16S rRNA sequencing
PCR-based identification
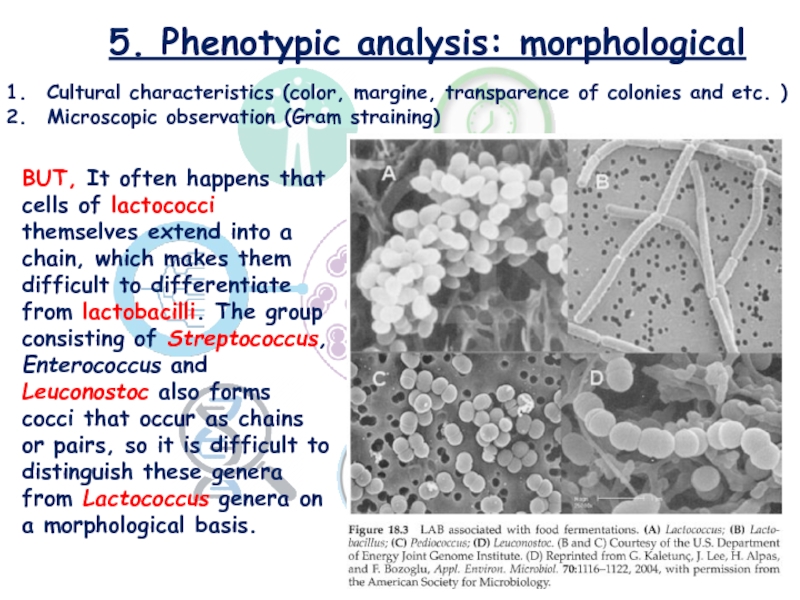
Слайд 175. Phenotypic analysis: morphological
Cultural characteristics (color, margine, transparence of colonies
and etc. )
Microscopic observation (Gram straining)
BUT, It often happens
that cells of lactococci themselves extend into a chain, which makes them difficult to differentiate from lactobacilli. The group consisting of Streptococcus, Enterococcus and Leuconostoc also forms cocci that occur as chains or pairs, so it is difficult to distinguish these genera from Lactococcus genera on a morphological basis.
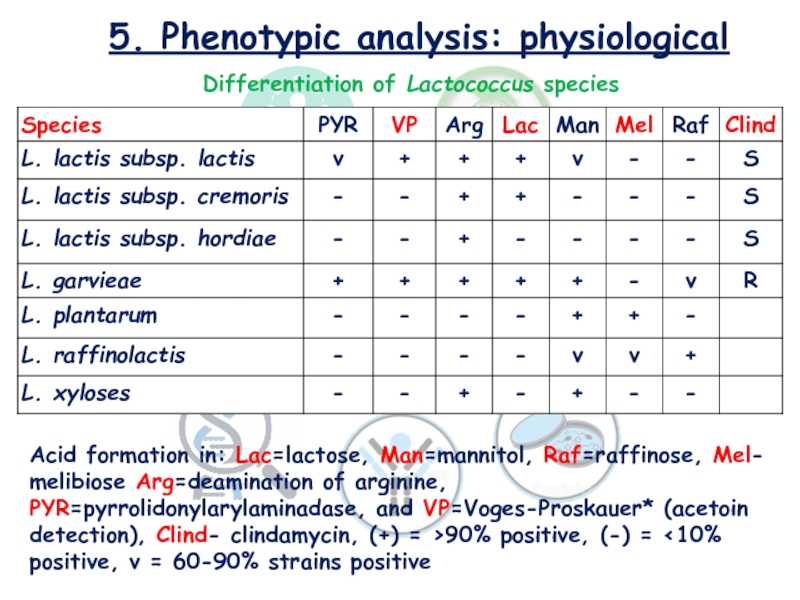
Слайд 185. Phenotypic analysis: physiological
Differentiation of Lactococcus species
Acid formation in: Lac=lactose, Man=mannitol, Raf=raffinose,
Mel- melibiose Arg=deamination of arginine, PYR=pyrrolidonylarylaminadase, and VP=Voges-Proskauer* (acetoin detection),
Clind- clindamycin, (+) = >90% positive, (-) = <10% positive, v = 60-90% strains positive
Слайд 196. Immunological (serological) analysis
Generally discovery of lactococcus as a separate
genera was investigated according to results of nucleic acid hybridization
studies and immunological relationships of superoxide dismutase. According Lancefield et al. representatives of Lactoccus sp. were classified into serological N group.
3. Serological
Slide agglutination, fluorescent
antibody staining, flow-cytometry based methods
Non-pathogenic
Pathogenic
Agglutination test for to proteolytic proteins (PepX, PepN, PepC, PepT, DIP and etc.) [Sakaki et al.]
Fish pathogen- L. garvieae (KG+ and KG type strains). KG+ type strain agglutinates with antiserum of KG 7409 strain, the KG type strain possesses a specific envelopelike substance, which inhibits agglutination with anti-KG 7409 serum.[Vendrel et al.]
Слайд 207. Genotypic techniques
Currently, there are two L.lactis ssp. cremoris and L. lactis ssp. lactis.
that have been sequenced for public release. This investigations has a
crucial role in understanding and manipulation of fermentation process to obtain desired product , also for fundamental researches of phylogenic diversity and epidemiology of pathogenic strains (L. garvieae ).
Methods
16S rRNA sequencing
(GTG)5-PCR fingerprinting
Genotyping IS elements (insertion elements)
Randomly amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR)
Multilocus sequence typing (for example 7 loci atpA, rpoA, pheS, pepN, bcaT, pepX, and 16S rRNA gene)
Слайд 217. Genotypic techniques: Genral scheme
Genotyping
16S rRNA sequencing
PCR
PCR reaction
Analysis of fragments
Sequencing of fragments
Analysis of sequences
Graph of relation
between isolates.
(GTG)5-PCR fingerprinting
PCR reaction (specific (GTG)5 region in genome
Analysis of fragments
Graph of relation between isolates
* So general mechanism is the same, the difference is in studying (genotyping) marker, the length and composition of which require designing specific primers and settling the reaction parameter. One more aspect that should be mentioned is the choose of marker for genotyping, cause the discriminatory index has a crucial role in diversification of isolates by strains.
Слайд 228. Manipulation (genetic or non-genetic)
Food industry (fermented products)
Agriculture (silage)
Antimicrobial
agents
Probiotics
Target therapy
Vaccine vector
Appli-cation
Слайд 238. Manipulation (genetic or non-genetic)
Food industry: cheese, butter, buttermilk, sour
cream and etc (non-genetic/genetic)
Agriculture, e.g. silage (non-genetic)
Source of antimicrobial
agents and preservatives called bacteriacines. For example “Nisin”. (non-genetic/ may include genetic manipulation for commercial overproduction)
Medicine- probiotics (non-genetic)
Medicine delivery factor (genetic [gene therapy])
New type of recombinant vaccine vector (genetic).
Слайд 248. Manipulation (genetic or non-genetic)
The food-grade bacterium Lactococcus lactis has
been extensively investigated during the last two decades as a
delivery vector for therapeutic proteins, DNA and vaccine antigens. The bacterium represents a safe, genetically tractable vector capable of producing heterologous therapeutic proteins at mucosal sites. Contributing this, recombinant L. lactis strains have been exploited as agents to treat inflammatory bowel disease, allergy and cancer.
Examples of vaccine delivery in practice – tetanus toxin C, pneumococcal diseases, staphylococcal enterotoxin B, H. pylory (HspA gene), and etc.
Слайд 268. Manipulation (genetic or non-genetic)
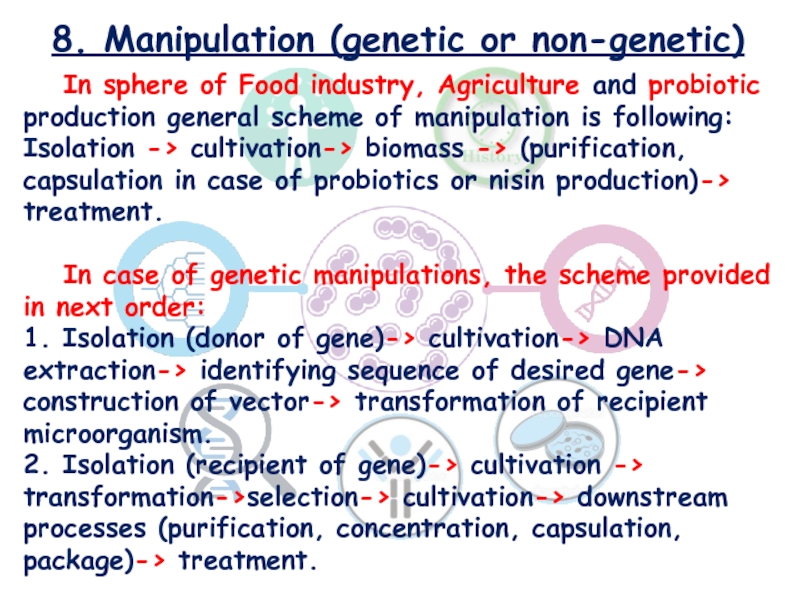
In sphere of Food industry, Agriculture
and probiotic production general scheme of manipulation is following: Isolation
-> cultivation-> biomass -> (purification, capsulation in case of probiotics or nisin production)-> treatment.
In case of genetic manipulations, the scheme provided in next order:
1. Isolation (donor of gene)-> cultivation-> DNA extraction-> identifying sequence of desired gene-> construction of vector-> transformation of recipient microorganism.
2. Isolation (recipient of gene)-> cultivation -> transformation->selection-> cultivation-> downstream processes (purification, concentration, capsulation, package)-> treatment.
Слайд 279. Facts
Lactoococcus sp. strains considered to be safe (GRAS) for
human and used in dairy product fermentation.
The genus contains strains
known to grow at or below 7˚C.
Today they are used extensively in food fermentations, which represent about 20% of the total economic value of fermented foods produced throughout the world.
The Lactococcus lactis ssp. lactis genome has 2 365 589 units (bp) of DNA, which contain 2 310 predicted genes. About 64% of the genes have assigned roles in the cell, while 20% match other hypothetical genes with unknown function. Almost 16% of the genes bear no resemblance to genes from other species and are considered to be unique to this bacterium.
Before 1985, representative of this genera were classified as Streptococcus and Enterococcus sp.
Слайд 28CONCLUSION
Description of the genus Lactococcus gen.nov. Lactococcus (lac.to.coc'cus, L.n.lac,
lactis milk., Gr.n.coccus, a grain or berry, M.L.masc.n. Lactococcus milk
coccus).
Spheres or ovoid cells occur singly, in pairs or in chains, and are often elongated in the direction of the chain. Gram-positive. Endospores are not formed. Non-motile. Not β-haemolytic. Facultatively anaerobic, catalase negative. Growth at 10°C but not at 45°C . Usually grows in 4 % (w/v) NaCL with the exception of L. lactis subsp. cremoris which only tolerates 2 % (w/v) NaCL. Chemoorganotrophs. Metabolism- fermentative. The predominant end product of glucose fermentation is L-lactic acid. Most strains react with group N antisera. Some strains possess low levels of menaquinones.
Слайд 29CONCLUSION
The major glycolipid of all strains is Glc(a1-2)Glc(a1-3)acyI2- Gro,
a constant minor component is Glc(a1-2), acyl- 6Glc(a1-3)acyI2Gro. All strains
contain phosphatidylglycerol and cardiolipin. Lipoteichoic acid structure and occurrence of aminophospholipids are species rather than genus-specific. Non-hydroxylated long-chain fatty acids are primarily of the straight-chain saturated and monounsaturated types; some strains produce cyclopropane-ring acids. The major fatty acids are hexadecanoic and cis- 11,12-octadecenoic acids; cis-11,12-methylenoctadecanoic acid is also present in major amounts in most strains with the exception of L. lactis subsp. hordniae and L. raffinolactis. The G+C content of the DNA ranges from 34 to 43 mol %.
Слайд 30CONCLUSION
Nucleic acid hybridization and comparative immunological studies demonstrate that
members of the genus Lactococcus are closely related to each
other but not to members of the genus Streptococcus or Enterococcus. Lactococci can be distinguished from streptococci and enterococci by their ability to grow at 10°C but not at 45°C. They are not β-haemolytic; some strains show a weak α-haemolytic reaction. They are non-motile.
Слайд 3110. References
D.Samaržija, N.Antunac, J.L. Havranek. Taxonomy, physiology and growth of
Lactococcus lactis: a review. Mljekarstvo 51 (1) 35-48, 2001.
K.H. Schleifer
et al. Transfer of Streptococcus lactis and Related Streptococci to the Genus Lactococcus gen. nov. System. Appl. Microbiol. 6, 183-195 (1985)
P. M. F. SHATTOCK AND A. T. R. MATTICK. THE SEROLOGICAL GROUPING OF STREPTOCOCCUS LACTIS (GROUP N) AND ITS RELATIONSHIP TO STREPTOCOCCUS FAECALIS .
J.M. Hardie and R.A. Whiley. Classification and overview of the genera Streptococcus and Enterococcus. Journal of Applied Microbiology Symposium Supplement 1997, 83, 1S–11S
Е.П. Мирошникова «Микробиология молока и молочных продуктов» 2005.
S. L. Cho et al. Lactococcus chungangensis sp. nov., a lactic acid bacterium isolated from activated sludge foam. International Journal of Systematic and Evolutionary Microbiology (2008), 58, 1844–1849
Montville et al. FOOD MICROBIOLOGY An Introduction. Second edition. Chapter 18. ASM Press American Society for Microbiology (2008)
P.M. Moraes et al. Comparison of phenotypic and molecular tests to identify lactic acid bacteria. Brazilian Journal of Microbiology 44, 1, 109-112 (2013)

Слайд 3210. References
9. M. Sakaki et al. Immunological and electrophoretic study
of the proteolytic enzymes from various Lactococcus and Lactobacillus strains.
Journal of Dairy Research (1995) 62 611-620
10. Vendrel et al. Lactococcus garvieae in fish: A review. Comparative Immunology, Microbiology & Infectious Diseases 29 (2006) 177–198
11. S. Altun et al. Genotyping of Lactococcus garvieae strains from rainbow trout (Oncorhynchus mykiss) by 16s rDNA sequencing. Bull. Eur. Ass. Fish Pathol., 24(2) 2004, 119
12. M. Nomura et al. Rapid PCR-Based Method Which Can Determine Both Phenotype and Genotype of Lactococcus lactis Subspecies. APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Vol. 68, No. 5, p. 2209–2213, May 2002
13. J. L. W. Rademaker et al. Diversity Analysis of Dairy and Nondairy Lactococcus lactis Isolates, Using a Novel Multilocus Sequence Analysis Scheme and (GTG)5-PCR Fingerprinting. APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Vol. 73, No. 22, p. 7128–7137, Nov. 2007
14. P. Nieto-Arribas et al. Genotypic and technological characterization of Lactococcus lactis isolates involved in processing of artisanal Manchego cheese. Journal of Applied Microbiology 107 (2009) 1505–1517
Слайд 3310. References
15. E. Ferna´ndez et al. Comparative Phenotypic and Molecular
Genetic Profiling of Wild Lactococcus lactis subsp. lactis Strains of
the L. lactis subsp. lactis and L. lactis subsp. cremoris Genotypes, Isolated from Starter-Free Cheeses Made of Raw Milk. APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Vol. 77, No. 15,p. 5324–5335,Aug. 2011
16. *I. Boucher et al. Novel Food-Grade Plasmid Vector Based on Melibiose Fermentation for the Genetic Engineering of Lactococcus lactis. APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Vol 68, No. 12, p. 6152–6161,Dec. 2002
17. L. G. Stoyanova et al. Isolation and Identification of New Nisin-producing Lactococcus lactis subsp. Lactis from Milk. Applied Biochemistry and Microbiology, 2006, Vol. 42, No. 5, pp. 492–499.
18. Bahey-El-Din M, Gahan CG, Griffin BT. Lactococcus lactis as a cell factory for delivery of therapeutic proteins. Current Gene Therapy. 2010 Feb;10(1):34-45.
19. K. Robinson et al. Mucosal and Cellular Immune Responses Elicited by Recombinant Lactococcus lactis Strains Expressing Tetanus Toxin Fragment C. INFECTION AND IMMUNITY, Vol 72, No. 5, p. 27532761,May 2004
20. J.V. Hernandez et al. Targeting diseases with genetically engineered Lactococcus lactis and its course towards medical translation. Expert Opin. Biol. Ther. (2011) 11(3):261-267

Слайд 3410. References
21. S. B. Hanniffy et al. Mucosal Delivery of
a Pneumococcal Vaccine Using Lactococcus lactis Affords Protection against Respiratory
Infection. The Journal of Infectious Diseases 2007; 195:185–93
22. G. F. Asensi et al. Oral immunization with Lactococcus lactis secreting attenuated recombinant staphylococcal enterotoxin B induces a protective immuneresponse in a murine model. Microbial Cell Factories 2013, 12:32
23. M. Medina et al. Lactococcus lactis as an adjuvant and delivery vehicle of antigens against pneumococcal respiratory infections. Bioengineered Bugs 1:5, 313-325; 2010
24. X.Z. Zhang et al. Expression of Helicobacter pylori hspA Gene in Lactococcus lactis NICE System and Experimental Study on Its Immunoreactivity. Hindawi Publishing Corporation Gastroenterology Research and Practice Volume 2015, Article ID 750932, 6 pages
25.Q. Gu et al. Oral vaccination ofmice againstHelicobacter pylori with recombinantLactococcus lactis expressing urease subunit B. FEMS Immunol Med Microbiol 56 (2009) 197–203
26. Bahey-El-Din M, Gahan CG, Griffin BT. Lactococcus lactis as a cell factory for delivery of therapeutic proteins. Current Gene Therapy. 2010 Feb;10(1):34-45.
Слайд 3510. References
Links:
1. https://en.wikipedia.org/wiki/Lactococcus
2. http://genome.jgi.doe.gov/laccr/laccr.home.html
3. http://textbookofbacteriology.net/lactics.html