Слайд 1Key Terms
The replicon is a unit of the genome in
which DNA is replicated. Replicons can be linear or circular.
Each replicon contains an origin for initiation of replication.
The origin is a sequence of DNA at which replication is initiated.
A terminus is a segment of DNA at which replication ends.
A replication eye is a region in which DNA has been replicated within a longer, unreplicated region.
A replication fork (Growing point) is the point at which strands of parental duplex DNA are separated so that replication can proceed. A complex of proteins including DNA polymerase is found at the fork.
Unidirectional replication refers to the movement of a single replication fork from a given origin.
Bidirectional replication describes a system in which an origin generates two
replication forks that proceed away from the origin in opposite directions.
S phase is the restricted part of the eukaryotic cell cycle during which synthesis of DNA occurs.
Key Concepts
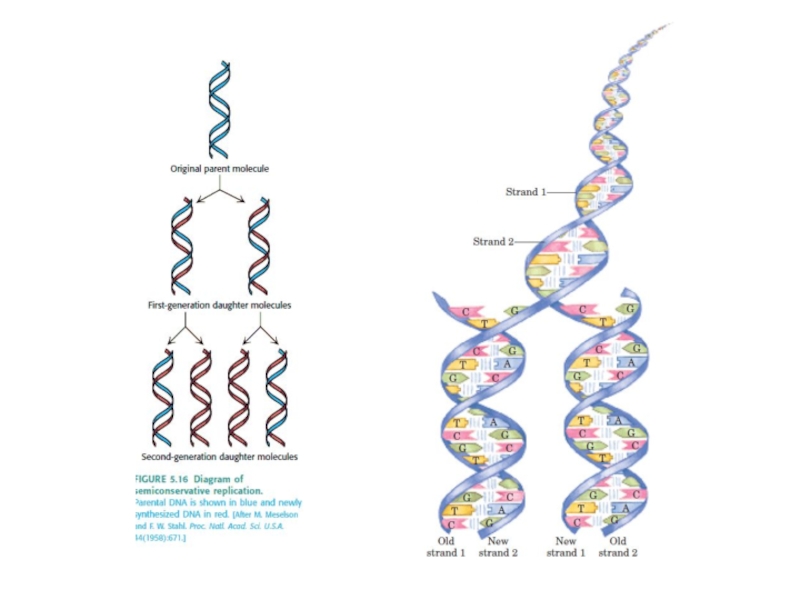
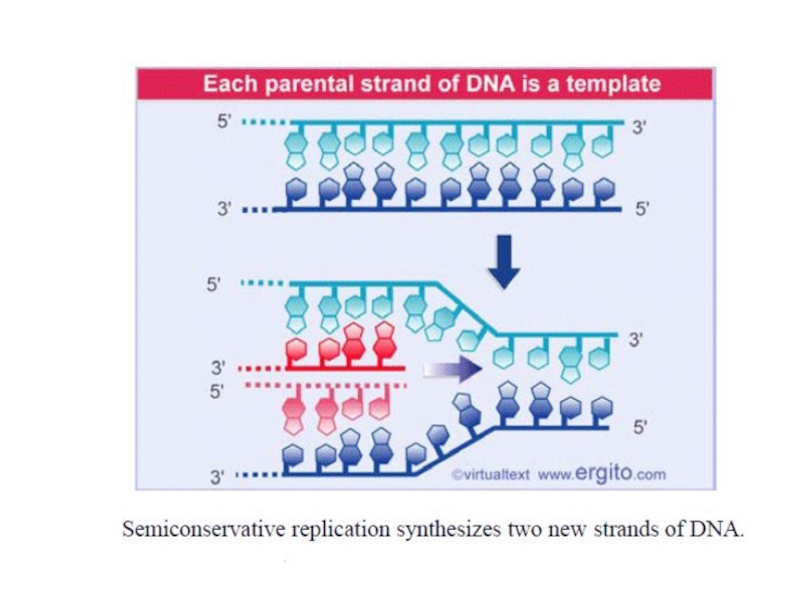
A replicated region appears as an eye within nonreplicated DNA.
A replication fork is initiated at the origin and then moves sequentially along DNA.
Replication is unidirectional when a single replication fork is created at an origin.
Eukaryotic replicons are 40-100 kb in length.
Each eukaryotic chromosome contains many replicons.
Individual replicons are activated at characteristic times during S phase.
Regional activation patterns suggest that replicons near one another are activated at the same time.

Слайд 4
Nick translation describes the ability of E. coli DNA polymerase
I to use a nick as a starting point from
which one strand of a duplex DNA can be degraded and replaced by resynthesis of new material; is used to introduce radioactively labeled nucleotides into DNA in vitro.
DNA polymerase I has a unique 5’–3’ exonuclease activity that can be combined with DNA synthesis to perform nick translation.
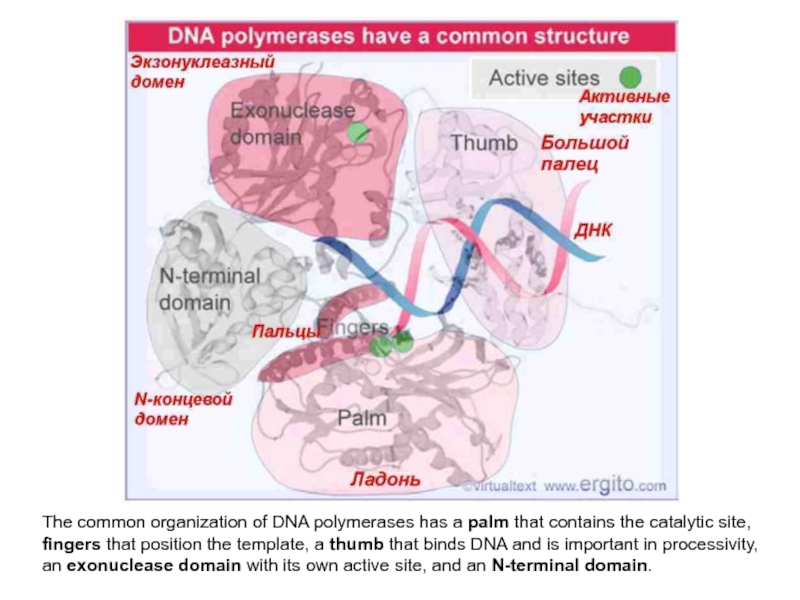
Слайд 5The common organization of DNA polymerases has a palm that
contains the catalytic site, fingers that position the template, a
thumb that binds DNA and is important in processivity, an exonuclease domain with its own active site, and an N-terminal domain.
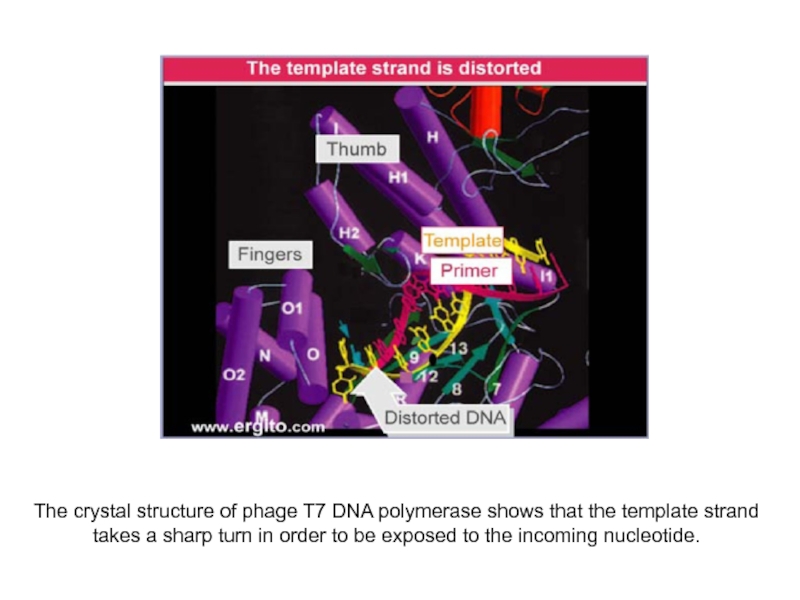
Слайд 6The crystal structure of phage T7 DNA polymerase shows that
the template strand takes a sharp turn in order to
be exposed to the incoming nucleotide.
Слайд 7
The leading strand of DNA is synthesized continuously in the
5’-3’ direction.
The lagging strand of DNA must grow overall in
the 5’-3’ direction and is synthesized discontinuously in the form of short fragments (5’-3’ ) that are later connected covalently.
Okazaki fragments are the short stretches of 1000-2000 bases produced during discontinuous replication; they are later joined into a covalently intact strand.
Semidiscontinuous replication is mode in which one new strand is synthesized continuously while the other is synthesized discontinuously.
Слайд 8
A helicase is an enzyme that uses energy provided by
ATP hydrolysis to separate the strands of a nucleic acid
duplex.
The single-strand binding protein (SSB) binds to single-stranded DNA, thereby preventing the DNA from forming a duplex.
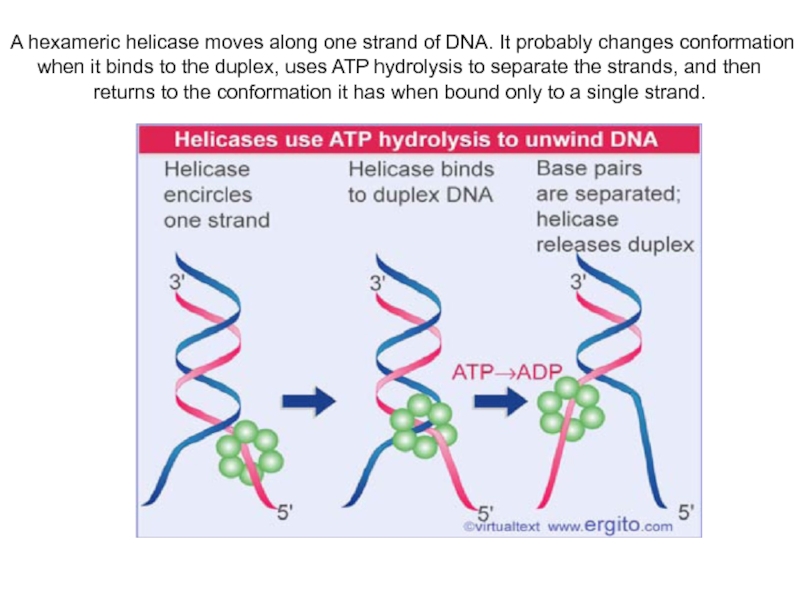
Слайд 9A hexameric helicase moves along one strand of DNA. It
probably changes conformation when it binds to the duplex, uses
ATP hydrolysis to separate the strands, and then returns to the conformation it has when bound only to a single strand.
Слайд 10Priming is required to start DNA synthesis
Key Terms
A primer is
a short sequence (often of RNA) that is paired with
one strand of DNA and provides a free 3’-OH end at which a DNA polymerase starts synthesis of a deoxyribonucleotide chain.
The primase is a type of RNA polymerase that synthesizes short segments of RNA that will be used as primers for DNA replication.
Key Concepts
All DNA polymerases require a 3’ –OH priming end to initiate DNA synthesis.
The priming end can be provided by an RNA primer, a nick in DNA, or a priming protein.
For DNA replication, a special RNA polymerase called a primase synthesizes an RNA chain that provides the priming end.
Priming of replication on double-stranded DNA always requires a replicase, SSB, and primase.
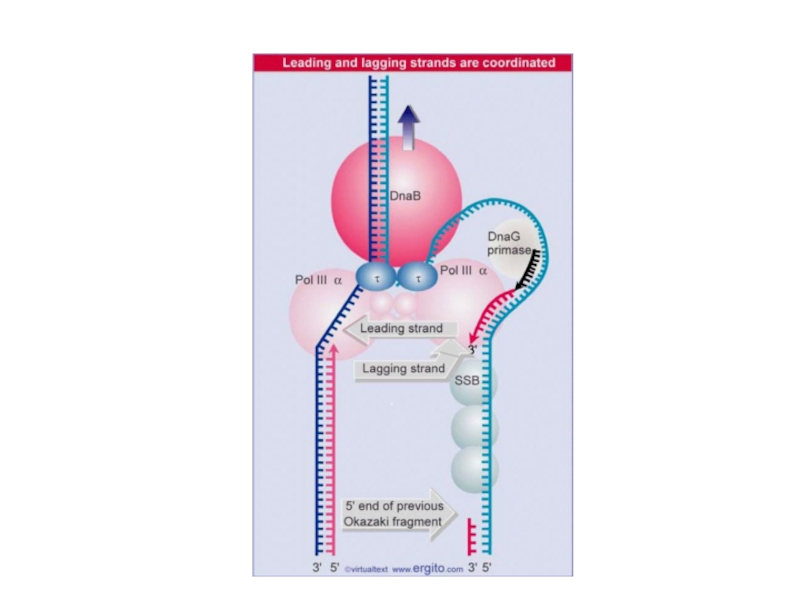
DnaB is the helicase that unwinds DNA for replication in E. coli.
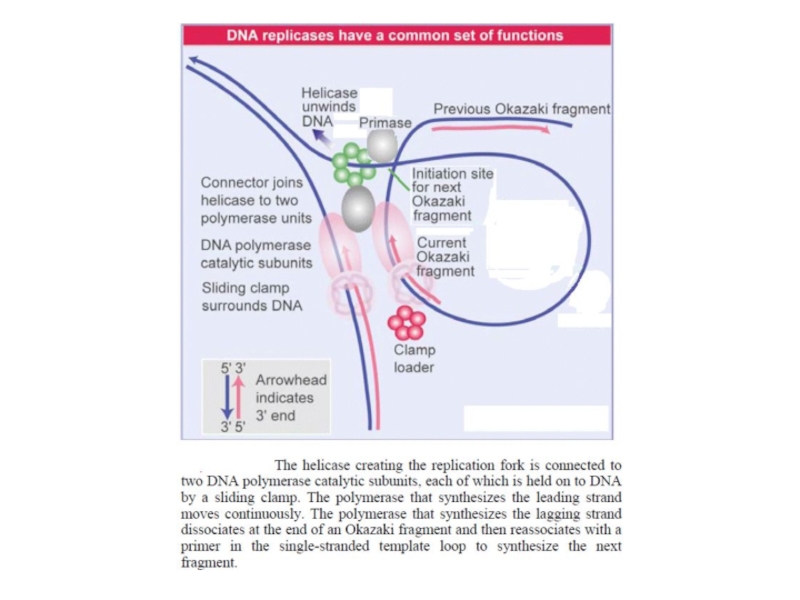
Слайд 11Coordinating synthesis of the lagging and leading strands
Key Concepts
Different enzyme
units are required to synthesize the leading and lagging strands.
In
E. coli both these units contain the same catalytic subunit (DnaE).
In other organisms, different catalytic subunits may be required for each strand.
DNA polymerase holoenzyme has 3 subcomplexes
The clamp loader is a 5 subunit protein complex which is responsible for loading the ß clamp on to DNA at the replication fork.
The E. coli replicase DNA polymerase III is a 900 kD complex with a dimeric structure.
Each monomeric unit has a catalytic core, a dimerization subunit, and a processivity component.
A clamp loader places the processivity subunits on DNA, and they form a circular clamp around the nucleic acid.
One catalytic core is associated with each template strand.
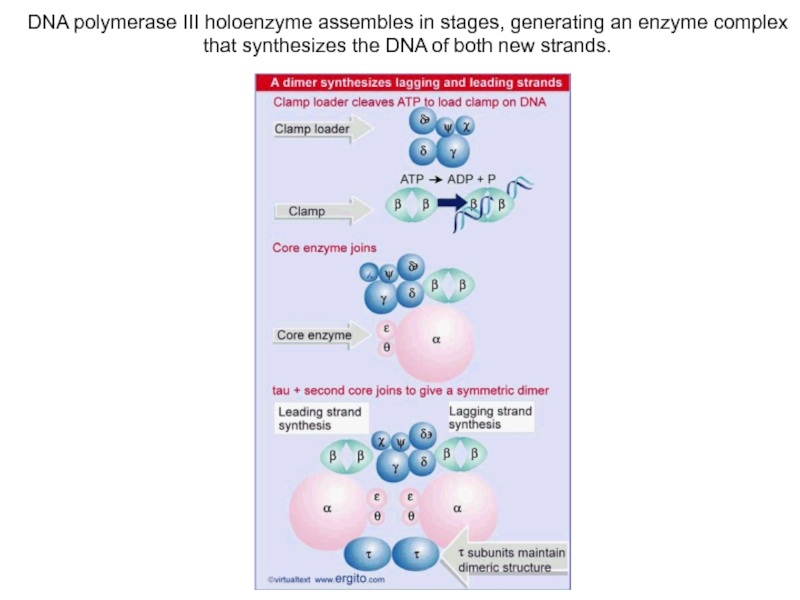
Слайд 12DNA polymerase III holoenzyme assembles in stages, generating an enzyme
complex that synthesizes the DNA of both new strands.
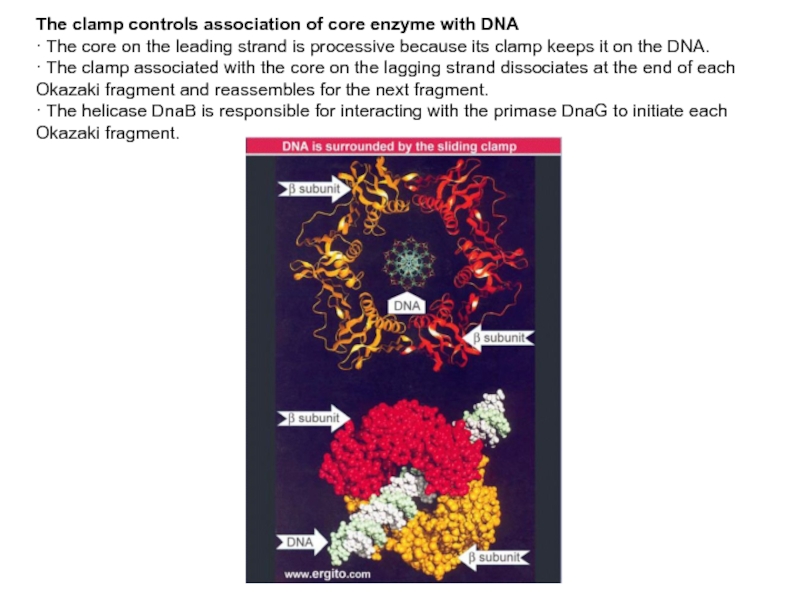
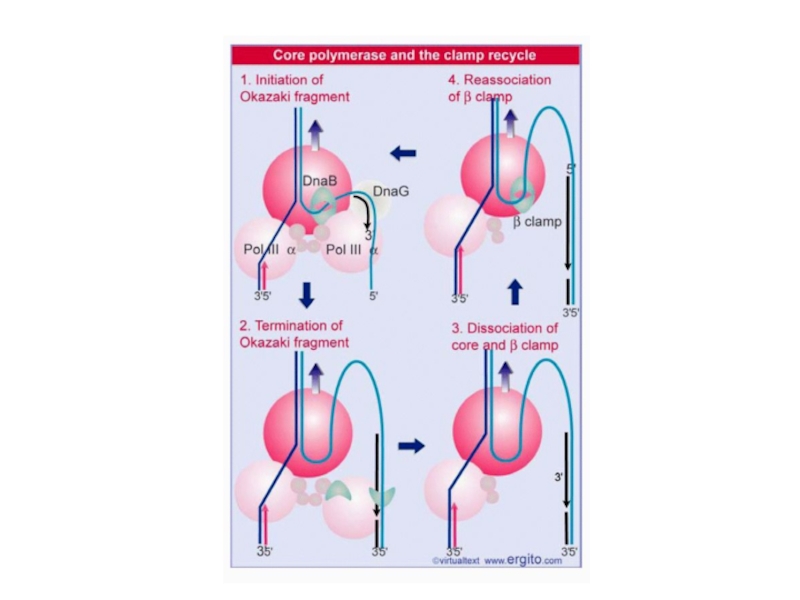
Слайд 13The clamp controls association of core enzyme with DNA
· The
core on the leading strand is processive because its clamp
keeps it on the DNA.
· The clamp associated with the core on the lagging strand dissociates at the end of each Okazaki fragment and reassembles for the next fragment.
· The helicase DnaB is responsible for interacting with the primase DnaG to initiate each Okazaki fragment.
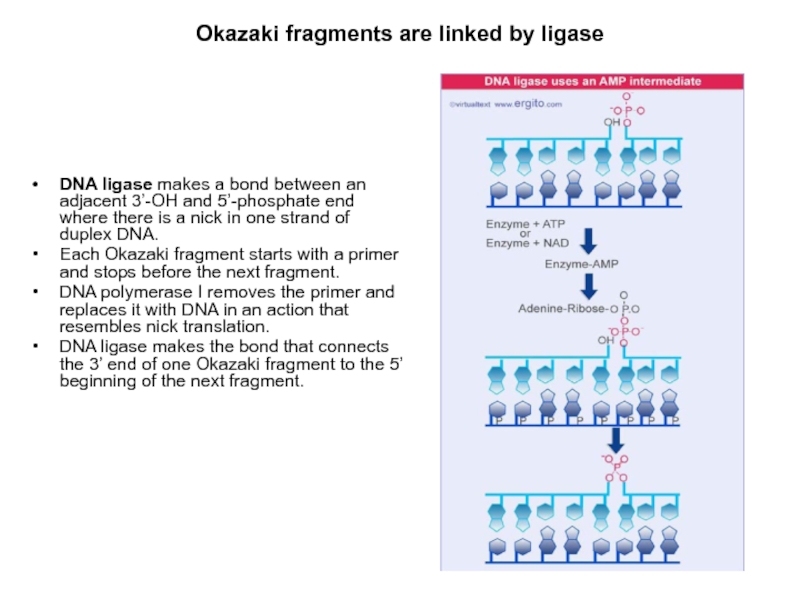
Слайд 17Okazaki fragments are linked by ligase
DNA ligase makes a bond
between an adjacent 3’-OH and 5’-phosphate end where there is
a nick in one strand of duplex DNA.
Each Okazaki fragment starts with a primer and stops before the next fragment.
DNA polymerase I removes the primer and replaces it with DNA in an action that resembles nick translation.
DNA ligase makes the bond that connects the 3’ end of one Okazaki fragment to the 5’ beginning of the next fragment.
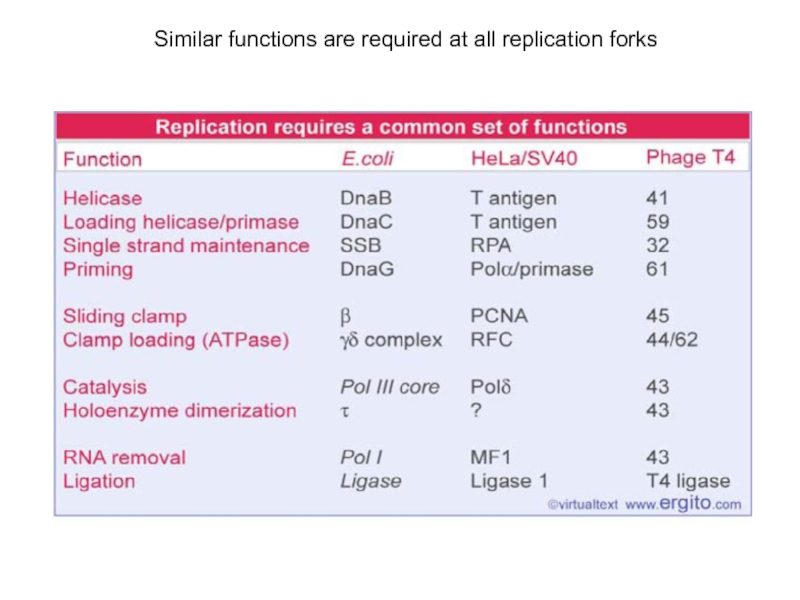
Слайд 18Similar functions are required at all replication forks
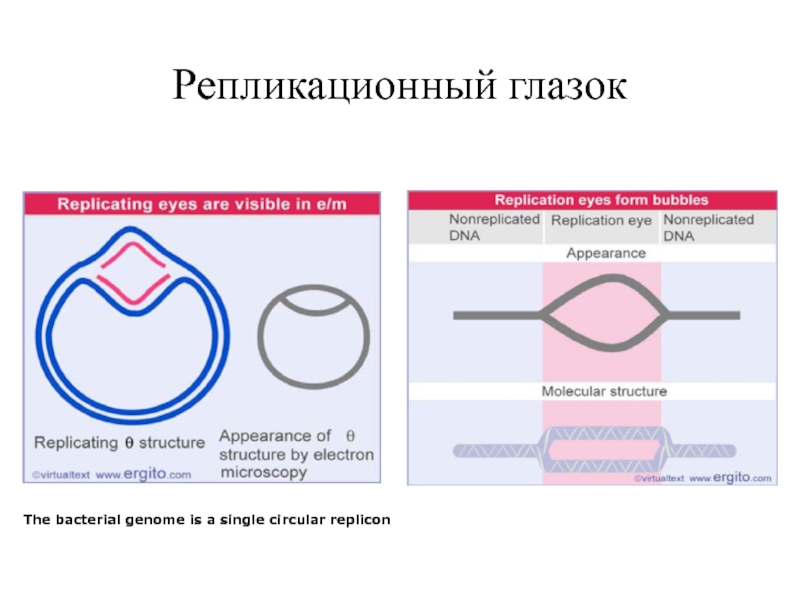
Слайд 19Репликационный глазок
The bacterial genome is a single circular replicon
Слайд 20Creating the replication forks at an origin
The origin of E.
coli,oriC, is 245 bp in length
Origins in S. cerevisiae are
short A·T-rich sequences that have an essential 11 bp sequence.
The ORC is a complex of 6 proteins that binds to an ARS.
Initiation at oriC requires the sequential assembly of a large protein complex.
DnaA binds to short repeated sequences and forms an oligomeric complex that melts DNA.
6 DnaC monomers bind each hexamer of DnaB and this complex binds to the origin.
A hexamer of DnaB forms the replication fork.
A short region of A·T-rich DNA is melted.
Gyrase and SSB are also required.
·
Слайд 22DNA REPLICATION Summary
DNA synthesis occurs by semidiscontinuous replication, in which
the leading strand of DNA growing 5 ¢ –3 ¢
is extended continuously, but the lagging strand that grows overall in the opposite 3 ¢ –5 ¢ direction is made as short Okazaki fragments, each synthesized 5 ¢ –3 ¢ . The leading strand and each Okazaki fragment of the lagging strand initiate with an RNA primer that is extended by DNA polymerase. Bacteria and eukaryotes each possess more than one DNA polymerase activity. DNA polymerase III synthesizes both lagging and leading strands in E. coli. Many proteins are required for DNA polymerase III action and several constitute part of the replisome within which it functions.
The replisome contains an asymmetric dimer of DNA polymerase III; each new DNA strand is synthesized by a different core complex containing a catalytic ( a) subunit. Processivity of the core complex is maintained by the b clamp, which forms a ring round DNA. The clamp is loaded on to DNA by the clamp loader complex. Clamp/clamp loader pairs with similar structural features are widely found in both prokaryotic and eukaryotic replication systems.
The looping model for the replication fork proposes that, as one half of the dimer advances to synthesize the leading strand, the other half of the dimer pulls DNA through as a single loop that provides the template for the lagging strand. The transition from completion of one Okazaki fragment to the start of the next requires the lagging strand catalytic subunit to dissociate from DNA and then to reattach to a b clamp at the priming site for the next Okazaki fragment. DnaB provides the helicase activity at a replication fork; this depends on ATP cleavage.
DnaB may function by itself in oriC replicons to provide primosome activity by interacting periodically with DnaG, which provides the primase that synthesizes RNA.
Phage T4 codes for a replication apparatus consisting of 7 proteins: DNA polymerase, helicase, single-strand binding protein, priming activities, and accessory proteins. Similar functions are required in other replication systems, including a HeLa cell system that replicates SV40 DNA. Different enzymes, DNA polymerase a and DNA polymerase d, initiate and elongate the new strands of DNA.
The fX priming event also requires DnaB, DnaC, and DnaT. PriA is the component that defines the primosome assembly site (pas) for fX replicons; it displaces SSB from DNA in an action that involves cleavage of ATP. PriB and PriC are additional components of the primosome. The importance of the primosome for the bacterial cell is that it is used to restart replication at forks that stall when they encounter damaged DNA.
The common mode of origin activation involves an initial limited melting of the double helix, followed by more general unwinding to create single strands. Several proteins act sequentially at the E. coli origin. Replication is initiated at oriC in E. Coli when DnaA binds to a series of 9 bp repeats. This is followed by binding to a series of 13 bp repeats, where it uses hydrolysis of ATP to generate the energy to separate the DNA strands. The pre-priming complex of DnaC-DnaB displaces DnaA. DnaC is released in a reaction that depends on ATP hydrolysis; DnaB is joined by the replicase enzyme, and replication is initiated by two forks that set out in opposite directions. Similar events occur at the lambda origin, where phage proteins O and P are the counterparts of bacterial proteins DnaA and DnaC, respectively. In SV40 replication, several of these activities are combined in the functions of T antigen.
The availability of DnaA at the origin is an important component of the system that determines when replication cycles should initiate. Following initiation of replication, DnaA hydrolyzes its ATP under the stimulus of the b sliding clamp, generating an inactive form of the protein. Also, oriC must compete with the dat site for binding DnaA.
Several sites that are methylated by the Dam methylase are present in the E. Coli origin, including those of the 13-mer binding sites for DnaA. The origin remains hemimethylated and is in a sequestered state for ~10 minutes following initiation of a replication cycle. During this period it is associated with the membrane, and reinitiation of replication is repressed. The protein SeqA is involved in sequestration and may interact with DnaA.
After cell division, nuclei of eukaryotic cells have a licensing factor that is needed to initiate replication. Its destruction after initiation of replication prevents further replication cycles from occurring in yeast. Licensing factor cannot be imported into the nucleus from the cytoplasm, and can be replaced only when the nuclear membrane breaks down during mitosis.
The origin is recognized by the ORC proteins, which in yeast remain bound throughout the cell cycle. The protein Cdc6 is available only at S phase. In yeast it is synthesized during S phase and rapidly degraded. In animal cells it is synthesized continuously, but is exported from the nucleus during S phase. The presence of Cdc6 allows the MCM proteins to bind to the origin. The MCM proteins are required for initiation. The action of Cdc6 and the MCM proteins provides the licensing function.