Разделы презентаций
- Разное
- Английский язык
- Астрономия
- Алгебра
- Биология
- География
- Геометрия
- Детские презентации
- Информатика
- История
- Литература
- Математика
- Медицина
- Менеджмент
- Музыка
- МХК
- Немецкий язык
- ОБЖ
- Обществознание
- Окружающий мир
- Педагогика
- Русский язык
- Технология
- Физика
- Философия
- Химия
- Шаблоны, картинки для презентаций
- Экология
- Экономика
- Юриспруденция
Forensic DNA Analysis
Содержание
- 1. Forensic DNA Analysis
- 2. Forensics, pertaining to the courts either criminal
- 3. Originally identification was limited to:Physical attributes such
- 4. Even though two unrelated humans differ in
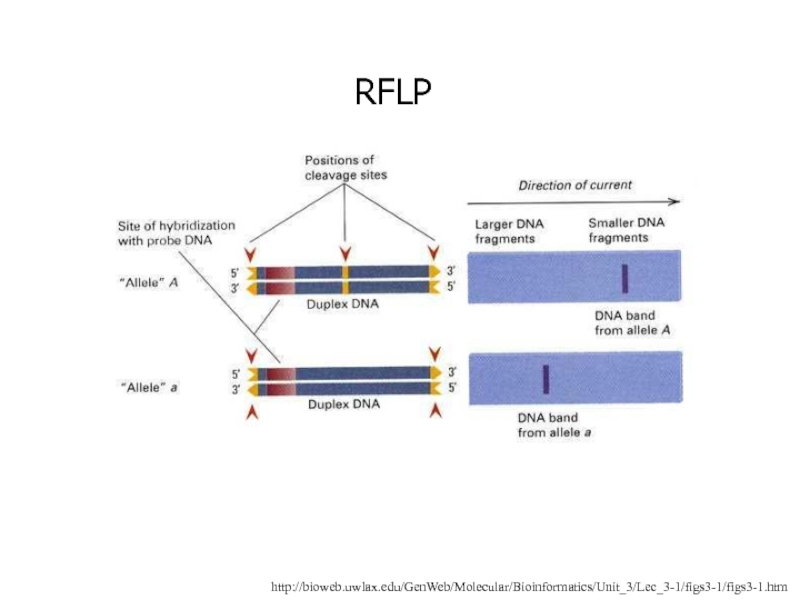
- 5. Restriction Fragment Length Polymorphism (RFLP)Detects a single
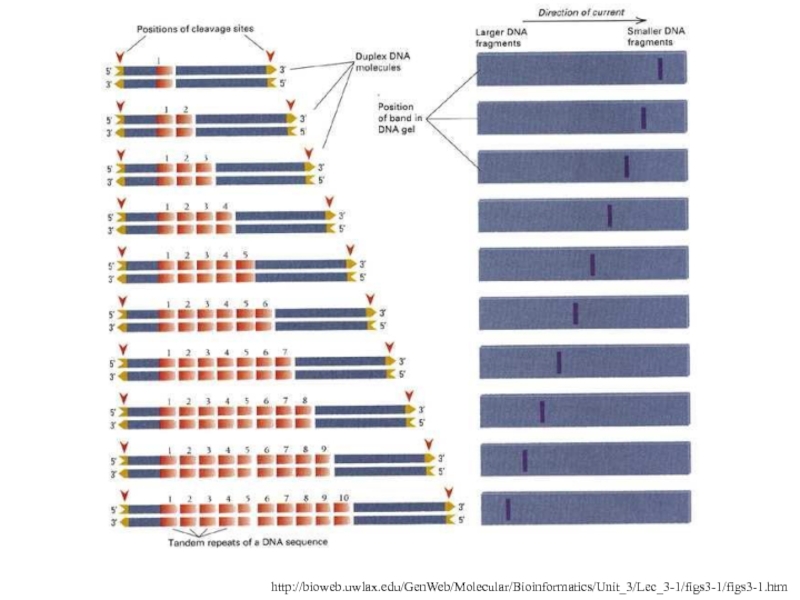
- 6. http://bioweb.uwlax.edu/GenWeb/Molecular/Bioinformatics/Unit_3/Lec_3-1/figs3-1/figs3-1.htmRFLP
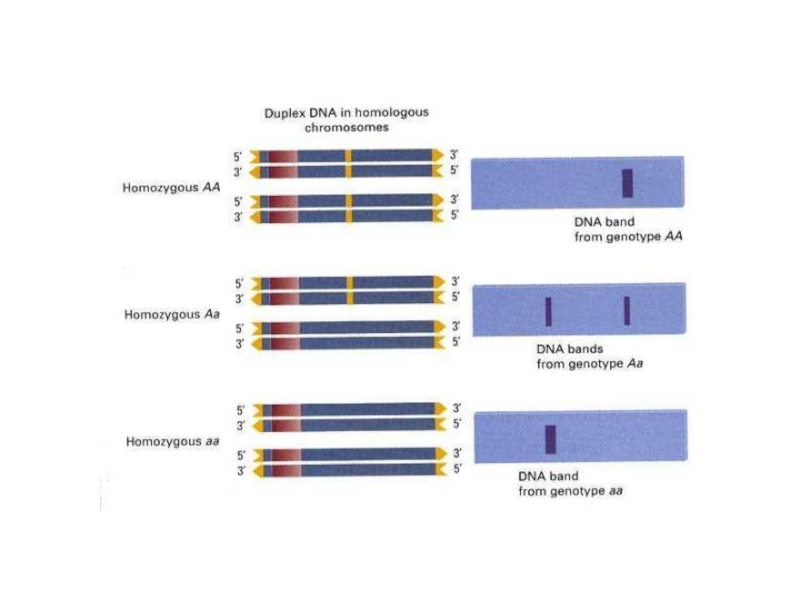
- 7. Слайд 7
- 8. DNA FingerprintingFirst described in 1985 by Alec
- 9. During the late 80s/early 90s US courts
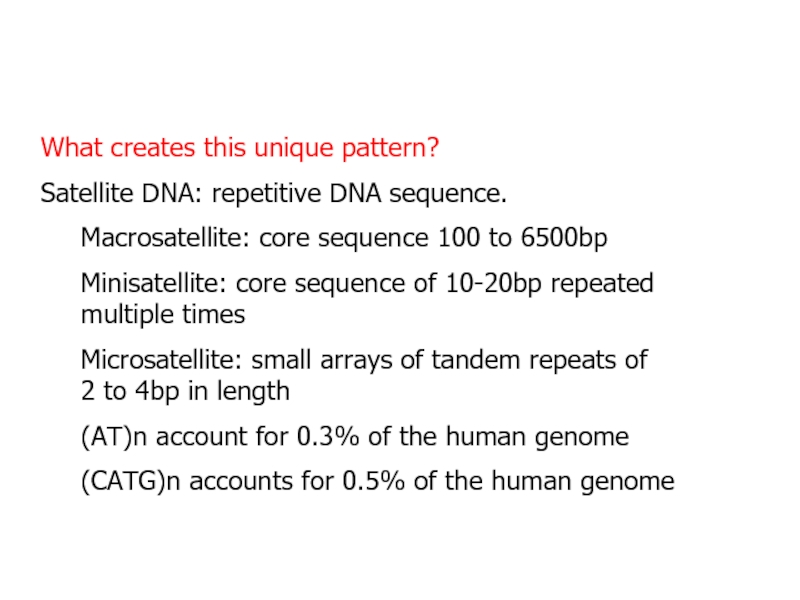
- 10. What creates this unique pattern?Satellite DNA: repetitive
- 11. Repeats of Satellite DNARepeat units vary in
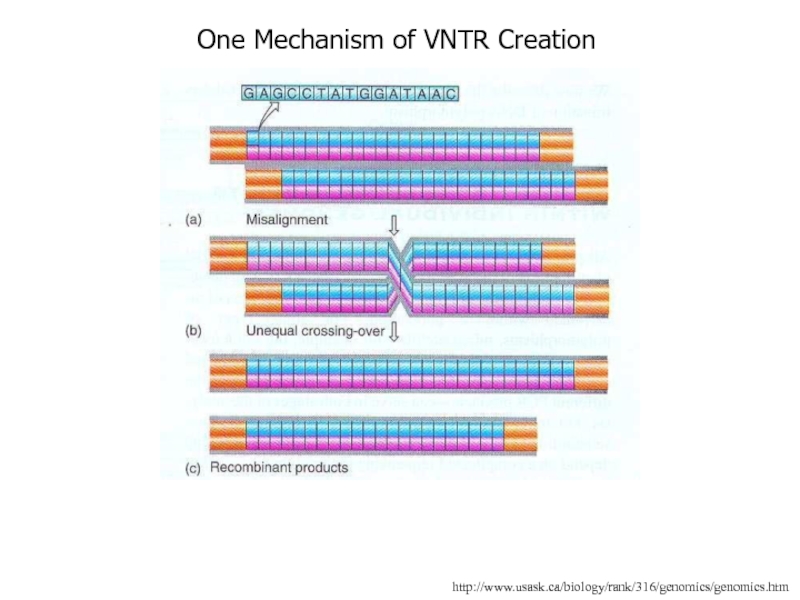
- 12. http://www.usask.ca/biology/rank/316/genomics/genomics.htmOne Mechanism of VNTR Creation
- 13. Southern blotting can be used to visualize
- 14. http://bioweb.uwlax.edu/GenWeb/Molecular/Bioinformatics/Unit_3/Lec_3-1/figs3-1/figs3-1.htm
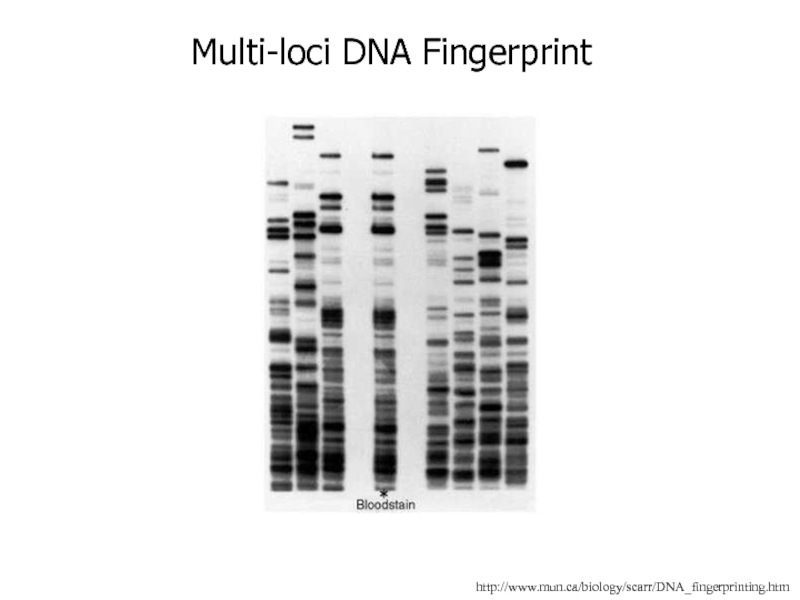
- 15. http://www.mun.ca/biology/scarr/DNA_fingerprinting.htmMulti-loci DNA Fingerprint
- 16. Multi-locus analysis of Dolly used to prove
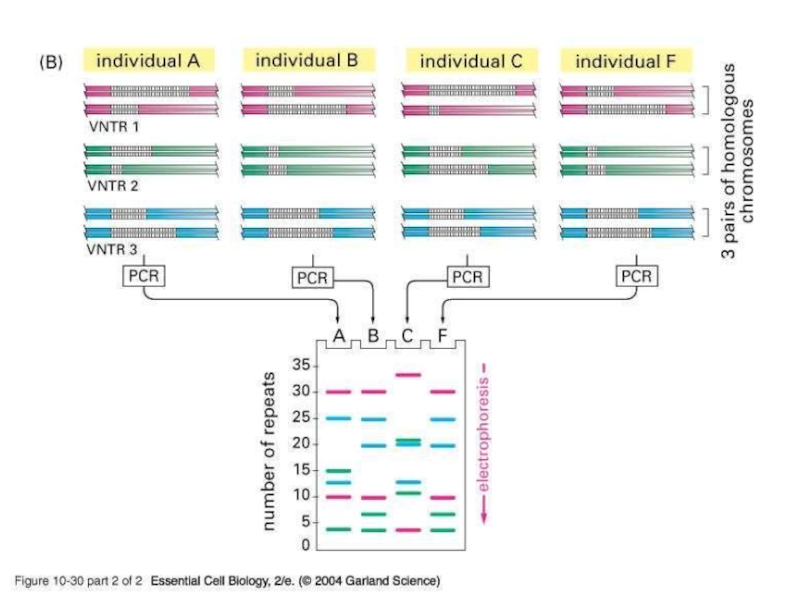
- 17. http://www.genelex.com/paternitytesting/paternityslide2.htmlSingle-Locus VNTRSingle-locus mini/microsatellite VNTRs generates at most
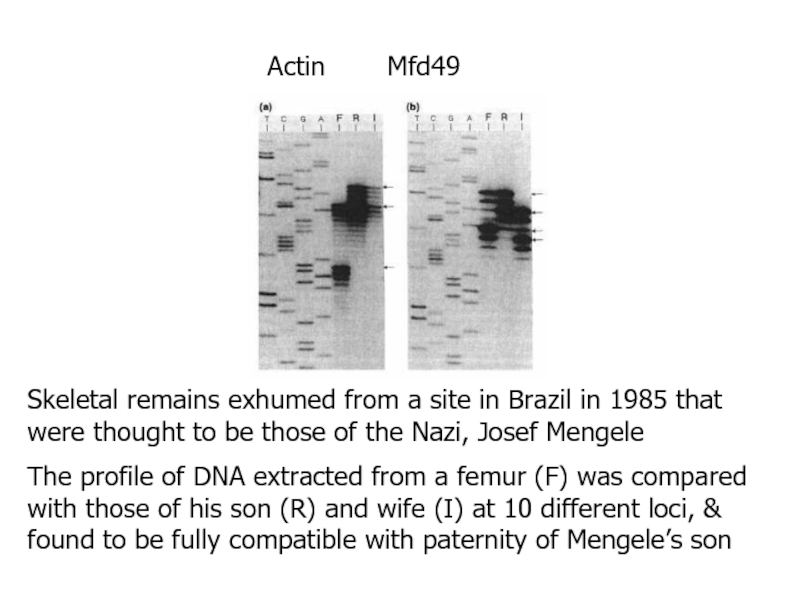
- 18. Skeletal remains exhumed from a site in
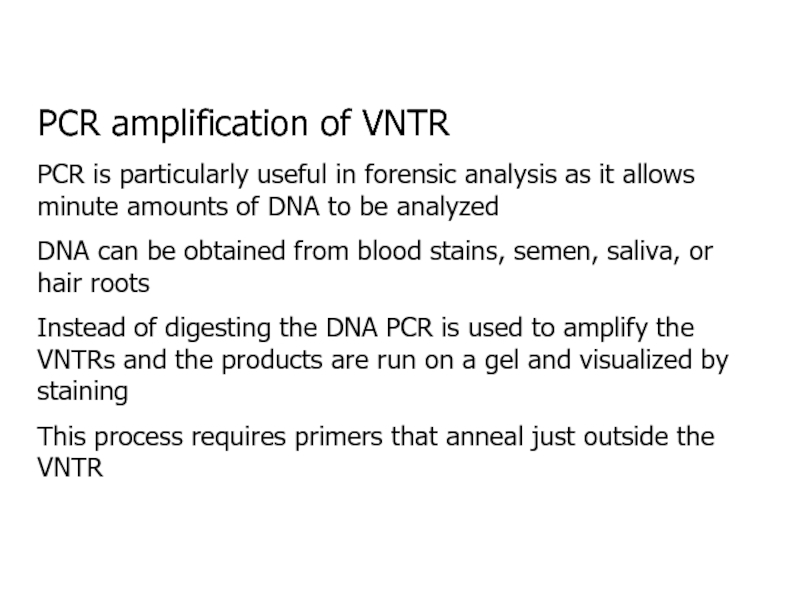
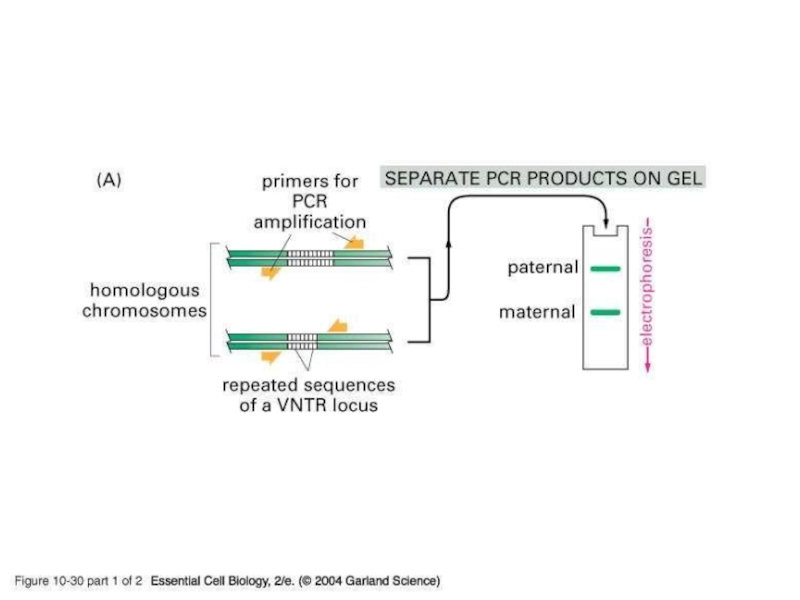
- 19. PCR amplification of VNTRPCR is particularly useful
- 20. Слайд 20
- 21. Слайд 21
- 22. Short Tandem Repeats (STR)Are a variation on
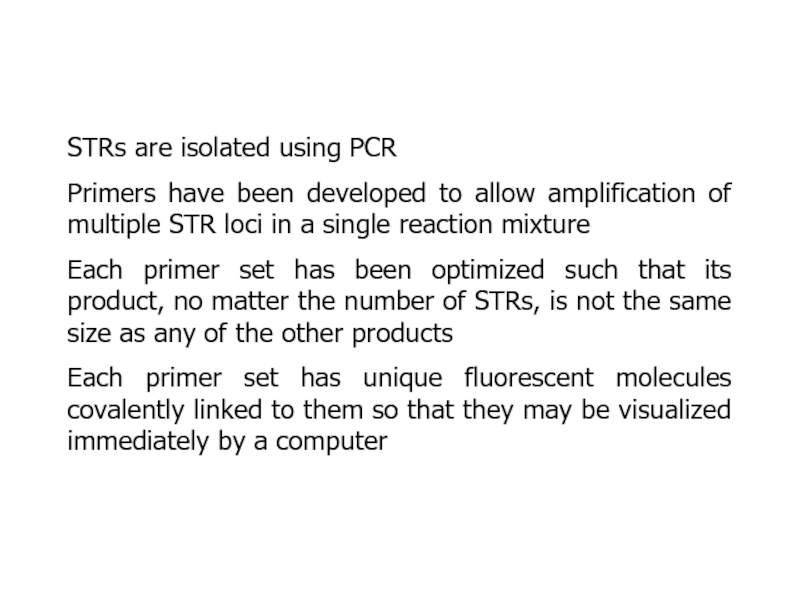
- 23. STRs are isolated using PCRPrimers have been
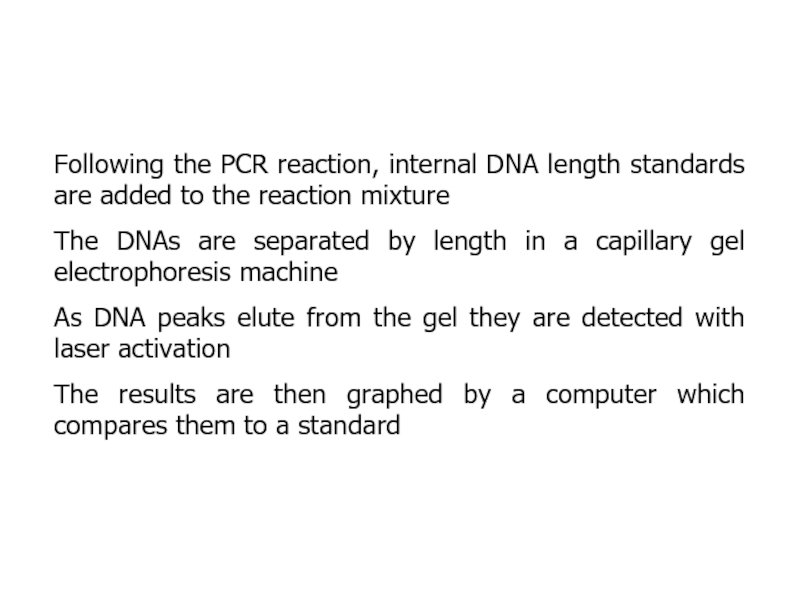
- 24. Following the PCR reaction, internal DNA length
- 25. http://www.biology.arizona.edu/human_bio/activities/blackett2/str_analysis.htmlAnalysis of 3 STRs, D3S1358, vWA, &
- 26. Example of a DNA profile using the
- 27. CODIS (Combined DNA Index System)In 1997, the
- 28. http://www.cstl.nist.gov/div831/strbase/images/codis.jpg
- 29. Newer Typing TechniquesMiniSTR uses shorter PCR primers
- 30. Скачать презентанцию
Forensics, pertaining to the courts either criminal or civilForensics DNA analysis is the use of DNA evidenceUsed in: paternity suites victim identification identifying suspects
Слайды и текст этой презентации
Слайд 2Forensics, pertaining to the courts either criminal or civil
Forensics DNA
analysis is the use of DNA evidence
suspectsСлайд 3Originally identification was limited to:
Physical attributes such as; ethnicity, gender,
height, weight, hair color, etc.
Friction-ridge identification or fingerprinting
Blood-antigen & serum
proteins, ABO blood groupsСлайд 4Even though two unrelated humans differ in their DNA only
by 0.1 to 0.2% there are still up to 6
million basepair differencesIt is these differences that are used to create a unique DNA “fingerprint” also known as DNA profile
Слайд 5Restriction Fragment Length Polymorphism (RFLP)
Detects a single basepair change in
DNA
Must occur within a restriction enzyme cleavage sequence to be
visibleOften used in disease screening such as in the detection of sickle cell anemia
DNA fragments are often visualized by Southern Blot
Слайд 6http://bioweb.uwlax.edu/GenWeb/Molecular/Bioinformatics/Unit_3/Lec_3-1/figs3-1/figs3-1.htm
RFLP
Слайд 8DNA Fingerprinting
First described in 1985 by Alec Jeffreys as a
method for identifying individuals by their unique pattern of DNA
bandingFirst use of DNA fingerprinting was in a 1985 immigration case in the UK. It identified a child as being the offspring of a British citizen
It was then used to rule out a suspect in a rape/murder case in England in 1986
Слайд 9During the late 80s/early 90s US courts questioned the validity
of DNA profiling
The debates centered on evidence collection procedures, training
of technicians, & the statistics used to establish a matchBy the mid 1990s DNA profiling was shown to be scientifically valid and DNA evidence became admissible
Слайд 10What creates this unique pattern?
Satellite DNA: repetitive DNA sequence.
Macrosatellite: core
sequence 100 to 6500bp
Minisatellite: core sequence of 10-20bp repeated multiple
timesMicrosatellite: small arrays of tandem repeats of 2 to 4bp in length
(AT)n account for 0.3% of the human genome
(CATG)n accounts for 0.5% of the human genome
Слайд 11Repeats of Satellite DNA
Repeat units vary in length from 2bp
to long stretches of 6000bp or more
These repeat units are
lined up head to tail and compose satellite DNA and are interspersed throughout the genomeThe number of units varies person to person
Thus these sequences are called VNTRs (variable number of tandem repeats)
A VNTR is a locus that is hypervariable due to a large number of alleles each characterized by a different number of repeat units
Слайд 13Southern blotting can be used to visualize the variation
Probes specific
to the repeat unit are hybridized to DNA cut with
a restriction enzyme that cuts just outside the VNTRThis allows for the difference in VNTR length to be detected
Two common probes are known are:
33.6 (AGGGCTGGAGG)18
31.5 (AGAGGTGGGCAGGTGG)29
These are multi-locus minisatellite probes and show about 17 different DNA bands for each individual
Слайд 16Multi-locus analysis of Dolly used to prove she was a
clone
1 –12 are control sheep
U is original udder cells
C is
cells from cultureD is Dolly blood cells
Слайд 17http://www.genelex.com/paternitytesting/paternityslide2.html
Single-Locus VNTR
Single-locus mini/microsatellite VNTRs generates at most two bands
Though not
as unique as multi-locus VNTRs they are simple to use
Multiple
single-locus VNTRs are used to give a DNA fingerprintСлайд 18Skeletal remains exhumed from a site in Brazil in 1985
that were thought to be those of the Nazi, Josef
MengeleThe profile of DNA extracted from a femur (F) was compared with those of his son (R) and wife (I) at 10 different loci, & found to be fully compatible with paternity of Mengele’s son
Actin Mfd49
Слайд 19PCR amplification of VNTR
PCR is particularly useful in forensic analysis
as it allows minute amounts of DNA to be analyzed
DNA
can be obtained from blood stains, semen, saliva, or hair rootsInstead of digesting the DNA PCR is used to amplify the VNTRs and the products are run on a gel and visualized by staining
This process requires primers that anneal just outside the VNTR
Слайд 22Short Tandem Repeats (STR)
Are a variation on VNTRs, but use
the smallest repeats units often only 2 to 4 bp
in lengthaatttttgtattttttttagagacggggtttcaccatgttggtcaggctgactatggagt
tattttaaggttaatatatataaagggtatgatagaacacttgtcatagtttagaacgaa
ctaacgatagatagatagatagatagatagatagatagatagatagatagatagacagat
tgatagtttttttttatctcactaaatagtctatagtaaacatttaattaccaatatttg
13 core loci of tetrameric repeats are tested together to make a DNA profile
The sequence above is locus D7S280 which is located on chromosome 7
Слайд 23STRs are isolated using PCR
Primers have been developed to allow
amplification of multiple STR loci in a single reaction mixture
Each
primer set has been optimized such that its product, no matter the number of STRs, is not the same size as any of the other productsEach primer set has unique fluorescent molecules covalently linked to them so that they may be visualized immediately by a computer
Слайд 24Following the PCR reaction, internal DNA length standards are added
to the reaction mixture
The DNAs are separated by length in
a capillary gel electrophoresis machineAs DNA peaks elute from the gel they are detected with laser activation
The results are then graphed by a computer which compares them to a standard
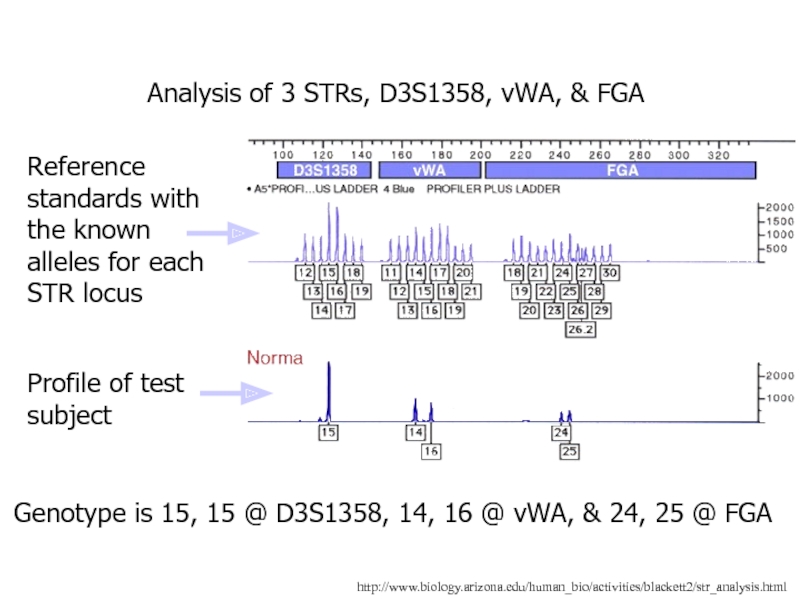
Слайд 25http://www.biology.arizona.edu/human_bio/activities/blackett2/str_analysis.html
Analysis of 3 STRs, D3S1358, vWA, & FGA
Reference standards with
the known alleles for each STR locus
Profile of test
subjectGenotype is 15, 15 @ D3S1358, 14, 16 @ vWA, & 24, 25 @ FGA
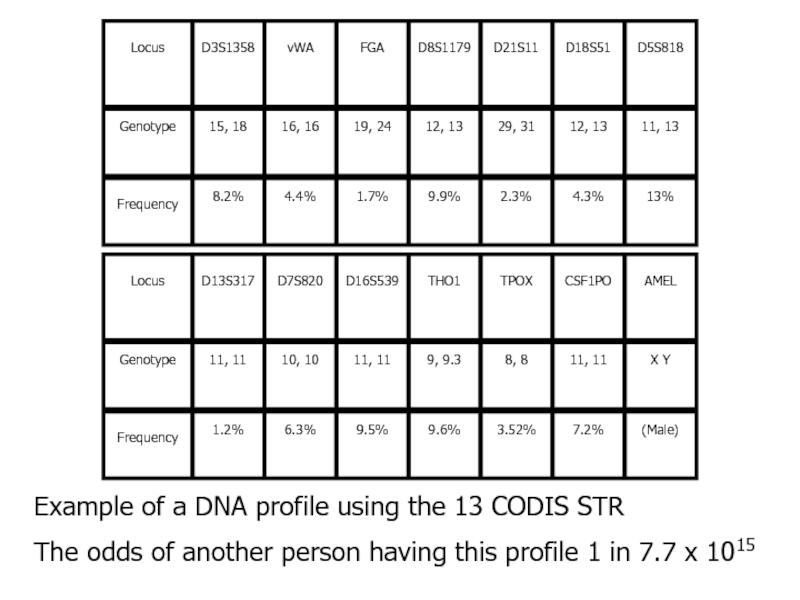
Слайд 26Example of a DNA profile using the 13 CODIS STR
The
odds of another person having this profile 1 in 7.7
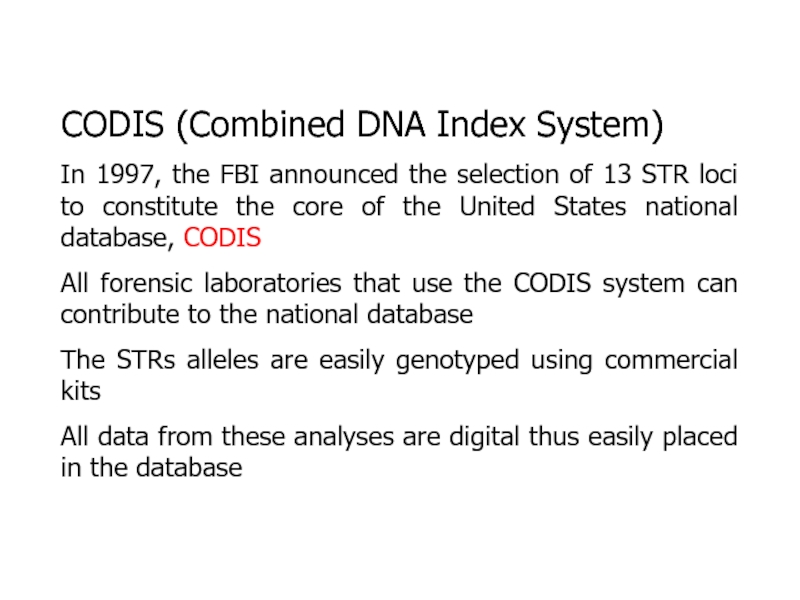
x 1015Слайд 27CODIS (Combined DNA Index System)
In 1997, the FBI announced the
selection of 13 STR loci to constitute the core of
the United States national database, CODISAll forensic laboratories that use the CODIS system can contribute to the national database
The STRs alleles are easily genotyped using commercial kits
All data from these analyses are digital thus easily placed in the database
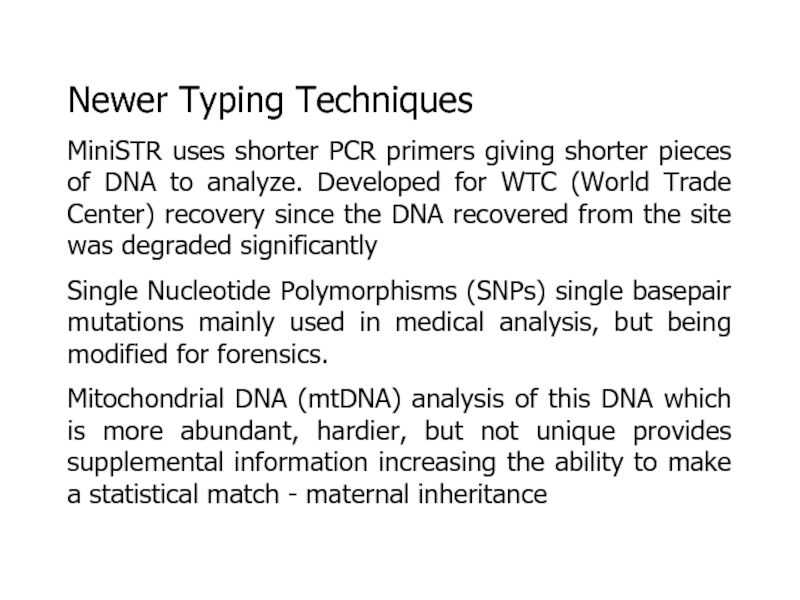
Слайд 29Newer Typing Techniques
MiniSTR uses shorter PCR primers giving shorter pieces
of DNA to analyze. Developed for WTC (World Trade Center)
recovery since the DNA recovered from the site was degraded significantlySingle Nucleotide Polymorphisms (SNPs) single basepair mutations mainly used in medical analysis, but being modified for forensics.
Mitochondrial DNA (mtDNA) analysis of this DNA which is more abundant, hardier, but not unique provides supplemental information increasing the ability to make a statistical match - maternal inheritance